The chromatin remodeler RSC prevents ectopic CENP-A propagation into pericentromeric heterochromatin at the chromatin boundary
- PMID: 36200823
- PMCID: PMC9638902
- DOI: 10.1093/nar/gkac827
The chromatin remodeler RSC prevents ectopic CENP-A propagation into pericentromeric heterochromatin at the chromatin boundary
Abstract
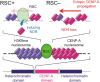
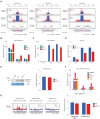
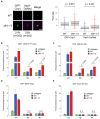
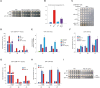
Centromeres of most eukaryotes consist of two distinct chromatin domains: a kinetochore domain, identified by the histone H3 variant, CENP-A, and a heterochromatic domain. How these two domains are separated is unclear. Here, we show that, in Schizosaccharomyces pombe, mutation of the chromatin remodeler RSC induced CENP-ACnp1 misloading at pericentromeric heterochromatin, resulting in the mis-assembly of kinetochore proteins and a defect in chromosome segregation. We find that RSC functions at the kinetochore boundary to prevent CENP-ACnp1 from spreading into neighbouring heterochromatin, where deacetylated histones provide an ideal environment for the spread of CENP-ACnp1. In addition, we show that RSC decompacts the chromatin structure at this boundary, and propose that this RSC-directed chromatin decompaction prevents mis-propagation of CENP-ACnp1 into pericentromeric heterochromatin. Our study provides an insight into how the distribution of distinct chromatin domains is established and maintained.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation.PLoS Genet. 2018 Aug 8;14(8):e1007572. doi: 10.1371/journal.pgen.1007572. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30089114 Free PMC article.
-
Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres.Science. 2008 Jan 4;319(5859):94-7. doi: 10.1126/science.1150944. Science. 2008. PMID: 18174443 Free PMC article.
-
Ectopic centromere nucleation by CENP--a in fission yeast.Genetics. 2014 Dec;198(4):1433-46. doi: 10.1534/genetics.114.171173. Epub 2014 Oct 7. Genetics. 2014. PMID: 25298518 Free PMC article.
-
Heterochromatin tells CENP-A where to go.Bioessays. 2008 Jun;30(6):526-9. doi: 10.1002/bies.20763. Bioessays. 2008. PMID: 18478529 Review.
-
The role of heterochromatin in centromere function.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):569-79. doi: 10.1098/rstb.2004.1611. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15905142 Free PMC article. Review.
Cited by
-
Epigenetic inheritance and boundary maintenance at human centromeres.Curr Opin Struct Biol. 2023 Oct;82:102694. doi: 10.1016/j.sbi.2023.102694. Epub 2023 Aug 30. Curr Opin Struct Biol. 2023. PMID: 37657353 Free PMC article. Review.
References
-
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Cleveland D.W.. Structural determinants for generating centromeric chromatin. Nature. 2004; 430:578–582. - PubMed
-
- Cheeseman I.M., Desai A.. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008; 9:33–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

