A caspase-RhoGEF axis contributes to the cell size threshold for apoptotic death in developing Caenorhabditis elegans
- PMID: 36201522
- PMCID: PMC9536578
- DOI: 10.1371/journal.pbio.3001786
A caspase-RhoGEF axis contributes to the cell size threshold for apoptotic death in developing Caenorhabditis elegans
Abstract
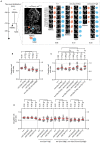
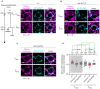
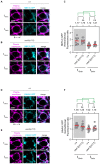
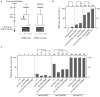
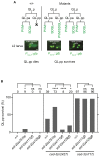
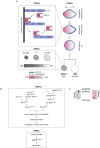
A cell's size affects the likelihood that it will die. But how is cell size controlled in this context and how does cell size impact commitment to the cell death fate? We present evidence that the caspase CED-3 interacts with the RhoGEF ECT-2 in Caenorhabditis elegans neuroblasts that generate "unwanted" cells. We propose that this interaction promotes polar actomyosin contractility, which leads to unequal neuroblast division and the generation of a daughter cell that is below the critical "lethal" size threshold. Furthermore, we find that hyperactivation of ECT-2 RhoGEF reduces the sizes of unwanted cells. Importantly, this suppresses the "cell death abnormal" phenotype caused by the partial loss of ced-3 caspase and therefore increases the likelihood that unwanted cells die. A putative null mutation of ced-3 caspase, however, is not suppressed, which indicates that cell size affects CED-3 caspase activation and/or activity. Therefore, we have uncovered novel sequential and reciprocal interactions between the apoptosis pathway and cell size that impact a cell's commitment to the cell death fate.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

