Identifying the genes impacted by cell proliferation in proteomics and transcriptomics studies
- PMID: 36201535
- PMCID: PMC9578628
- DOI: 10.1371/journal.pcbi.1010604
Identifying the genes impacted by cell proliferation in proteomics and transcriptomics studies
Abstract
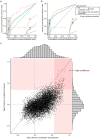
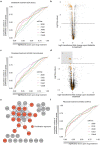
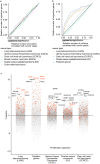
Hypothesis-free high-throughput profiling allows relative quantification of thousands of proteins or transcripts across samples and thereby identification of differentially expressed genes. It is used in many biological contexts to characterize differences between cell lines and tissues, identify drug mode of action or drivers of drug resistance, among others. Changes in gene expression can also be due to confounding factors that were not accounted for in the experimental plan, such as change in cell proliferation. We combined the analysis of 1,076 and 1,040 cell lines in five proteomics and three transcriptomics data sets to identify 157 genes that correlate with cell proliferation rates. These include actors in DNA replication and mitosis, and genes periodically expressed during the cell cycle. This signature of cell proliferation is a valuable resource when analyzing high-throughput data showing changes in proliferation across conditions. We show how to use this resource to help in interpretation of in vitro drug screens and tumor samples. It informs on differences of cell proliferation rates between conditions where such information is not directly available. The signature genes also highlight which hits in a screen may be due to proliferation changes; this can either contribute to biological interpretation or help focus on experiment-specific regulation events otherwise buried in the statistical analysis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

