Transcriptional changes in Plasmodium falciparum upon conditional knock down of mitochondrial ribosomal proteins RSM22 and L23
- PMID: 36201550
- PMCID: PMC9536634
- DOI: 10.1371/journal.pone.0274993
Transcriptional changes in Plasmodium falciparum upon conditional knock down of mitochondrial ribosomal proteins RSM22 and L23
Abstract
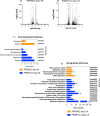
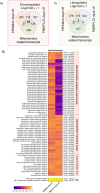
The mitochondrion of malaria parasites is an attractive antimalarial drug target, which require mitoribosomes to translate genes encoded in the mitochondrial (mt) DNA. Plasmodium mitoribosomes are composed of highly fragmented ribosomal RNA (rRNA) encoded in the mtDNA. All mitoribosomal proteins (MRPs) and other assembly factors are encoded in the nuclear genome. Here, we have studied one putative assembly factor, RSM22 (Pf3D7_1027200) and one large subunit (LSU) MRP, L23 (Pf3D7_1239100) in Plasmodium falciparum. We show that both proteins localize to the mitochondrion. Conditional knock down (KD) of PfRSM22 or PfMRPL23 leads to reduced cytochrome bc1 complex activity and increased sensitivity to bc1 inhibitors such as atovaquone and ELQ-300. Using RNA sequencing as a tool, we reveal the transcriptomic changes of nuclear and mitochondrial genomes upon KD of these two proteins. In the early phase of KD, while most mt rRNAs and transcripts of putative MRPs were downregulated in the absence of PfRSM22, many mt rRNAs and several MRPs were upregulated after KD of PfMRPL23. The contrast effects in the early phase of KD likely suggests non-redundant roles of PfRSM22 and PfMRPL23 in the assembly of P. falciparum mitoribosomes. At the late time points of KD, loss of PfRSM22 and PfMRPL23 caused defects in many essential metabolic pathways and transcripts related to essential mitochondrial functions, leading to parasite death. In addition, we enlist mitochondrial proteins of unknown function that are likely novel Plasmodium MRPs based on their structural similarity to known MRPs as well as their expression profiles in KD parasites.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

