CRISPR Spherical Nucleic Acids
- PMID: 36201634
- PMCID: PMC10430604
- DOI: 10.1021/jacs.2c07913
CRISPR Spherical Nucleic Acids
Abstract
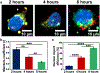
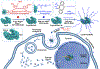
The use of CRISPR/Cas9 systems in genome editing has been limited by the inability to efficiently deliver the key editing components to and across tissues and cell membranes, respectively. Spherical nucleic acids (SNAs) are nanostructures that provide privileged access to both but have yet to be explored as a means of facilitating gene editing. Herein, a new class of CRISPR SNAs are designed and evaluated in the context of genome editing. Specifically, Cas9 ProSNAs comprised of Cas9 cores densely modified with DNA on their exteriors and preloaded with single-guide RNA were synthesized and evaluated for their genome editing capabilities in the context of multiple cell lines. The radial orientation of the DNA on the Cas9 protein surface enhances cellular uptake, without the need for electroporation or transfection agents. In addition, the Cas9 proteins defining the cores of the ProSNAs were fused with GALA peptides on their N-termini and nuclear localization signals on their C-termini to facilitate endosomal escape and maximize nuclear localization and editing efficiency, respectively. These constructs were stable against protease digestion under conditions that fully degrade the Cas9 protein, when not transformed into an SNA, and used to achieve genome editing efficiency between 32 and 47%. Taken together, these novel constructs and advances point toward a way of significantly broadening the scope of use and impact of CRISPR-Cas9 genome editing systems.
Figures


Similar articles
-
Hairpin Internal Nuclear Localization Signals in CRISPR-Cas9 Enhance Editing in Primary Human Lymphocytes.CRISPR J. 2025 Apr;8(2):105-119. doi: 10.1089/crispr.2024.0080. Epub 2025 Mar 31. CRISPR J. 2025. PMID: 40163415
-
Electroporation-Based CRISPR/Cas9 Gene Editing Using Cas9 Protein and Chemically Modified sgRNAs.Methods Mol Biol. 2019;1961:127-134. doi: 10.1007/978-1-4939-9170-9_9. Methods Mol Biol. 2019. PMID: 30912044
-
Regulating CRISPR/Cas9 Function through Conditional Guide RNA Control.Chembiochem. 2021 Jan 5;22(1):63-72. doi: 10.1002/cbic.202000423. Epub 2020 Nov 17. Chembiochem. 2021. PMID: 32833316 Free PMC article. Review.
-
Exploring the Cytoplasmic Retention of CRISPR-Cas9 in Eukaryotic Cells: The Role of Nuclear Localization Signals and Ribosomal Interactions.CRISPR J. 2025 Apr;8(2):120-136. doi: 10.1089/crispr.2024.0074. Epub 2025 Feb 28. CRISPR J. 2025. PMID: 40019800
-
CRISPR-Cas systems: ushering in the new genome editing era.Bioengineered. 2018;9(1):214-221. doi: 10.1080/21655979.2018.1470720. Bioengineered. 2018. PMID: 29968520 Free PMC article. Review.
Cited by
-
Distinct structural interactions of polyadenine and polythymine on gold nanoparticles: from single strands to duplexes.Chem Sci. 2025 Jul 31. doi: 10.1039/d5sc04459f. Online ahead of print. Chem Sci. 2025. PMID: 40787213 Free PMC article.
-
Spherical nucleic acids-based nanoplatforms for tumor precision medicine and immunotherapy.Mater Today Bio. 2023 Jul 26;22:100750. doi: 10.1016/j.mtbio.2023.100750. eCollection 2023 Oct. Mater Today Bio. 2023. PMID: 37545568 Free PMC article. Review.
-
Cellular Delivery of Large Functional Proteins and Protein-Nucleic Acid Constructs via Localized Electroporation.Nano Lett. 2023 Apr 26;23(8):3653-3660. doi: 10.1021/acs.nanolett.2c04374. Epub 2023 Feb 27. Nano Lett. 2023. PMID: 36848135 Free PMC article.
-
Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR-Cas9 ribonucleoprotein.Nat Biotechnol. 2024 Oct 16:10.1038/s41587-024-02437-3. doi: 10.1038/s41587-024-02437-3. Online ahead of print. Nat Biotechnol. 2024. PMID: 39415058
-
CRISPR/Cas-mediated "one to more" lighting-up nucleic acid detection using aggregation-induced emission luminogens.Nat Commun. 2024 Oct 3;15(1):8560. doi: 10.1038/s41467-024-52931-0. Nat Commun. 2024. PMID: 39362874 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

