Antibiotics and probiotics impact gut antimicrobial resistance gene reservoir in COVID-19 patients
- PMID: 36201636
- PMCID: PMC9543044
- DOI: 10.1080/19490976.2022.2128603
Antibiotics and probiotics impact gut antimicrobial resistance gene reservoir in COVID-19 patients
Abstract
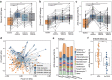
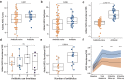
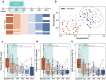
Dysbiosis of gut microbiota is well-described in patients with coronavirus 2019 (COVID-19), but the dynamics of antimicrobial resistance genes (ARGs) reservoir, known as resistome, is less known. Here, we performed longitudinal fecal metagenomic profiling of 142 patients with COVID-19, characterized the dynamics of resistome from diagnosis to 6 months after viral clearance, and reported the impact of antibiotics or probiotics on the ARGs reservoir. Antibiotic-naive patients with COVID-19 showed increased abundance and types, and higher prevalence of ARGs compared with non-COVID-19 controls at baseline. Expansion in resistome was mainly driven by tetracycline, vancomycin, and multidrug-resistant genes and persisted for at least 6 months after clearance of SARS-CoV-2. Patients with expanded resistome exhibited increased prevalence of Klebsiella sp. and post-acute COVID-19 syndrome. Antibiotic treatment resulted in further increased abundance of ARGs whilst oral probiotics (synbiotic formula, SIM01) significantly reduced the ARGs reservoir in the gut microbiota of COVID-19 patients during the acute infection and recovery phase. Collectively, these findings shed new insights on the dynamic of ARGs reservoir in COVID-19 patients and the potential role of microbiota-directed therapies in reducing the burden of accumulated ARGs.
Keywords: COVID-19; SIM01; antimicrobial resistance gene; gut microbiome; synbiotic formula.
Conflict of interest statement
FKLC and SCN are the scientific co-founders and sit on the board of Directors of GenieBiome Ltd. SCN, FKLC and ZX are inventors of patent applications related to SIM01. SCN has served as an advisory board member for Pfizer, Ferring, Janssen, and Abbvie and a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie, and Takeda. She has received research grants from Olympus, Ferring, and Abbvie. FKLC has served as an advisor and lecture speaker for Eisai Co. Ltd., AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co., and Takeda (China) Holdings Co. Ltd. All other co-authors have no conflict of interest.
Figures



References
-
- Resistance, I.C.G.o.A . 2019. No time to Wait. Securing the future from drug-resistant infections.
-
- Montassier E, Valdés-Mas R, Batard E, Zmora N, Dori-Bachash M, Suez J, Elinav E, et al. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat Microbiol. 2021;6:1043–1054. doi: 10.1038/s41564-021-00920-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
