Genetic subgroups inform on pathobiology in adult and pediatric Burkitt lymphoma
- PMID: 36201743
- PMCID: PMC10023728
- DOI: 10.1182/blood.2022016534
Genetic subgroups inform on pathobiology in adult and pediatric Burkitt lymphoma
Abstract
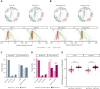
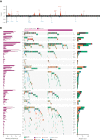
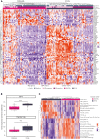
Burkitt lymphoma (BL) accounts for most pediatric non-Hodgkin lymphomas, being less common but significantly more lethal when diagnosed in adults. Much of the knowledge of the genetics of BL thus far has originated from the study of pediatric BL (pBL), leaving its relationship to adult BL (aBL) and other adult lymphomas not fully explored. We sought to more thoroughly identify the somatic changes that underlie lymphomagenesis in aBL and any molecular features that associate with clinical disparities within and between pBL and aBL. Through comprehensive whole-genome sequencing of 230 BL and 295 diffuse large B-cell lymphoma (DLBCL) tumors, we identified additional significantly mutated genes, including more genetic features that associate with tumor Epstein-Barr virus status, and unraveled new distinct subgroupings within BL and DLBCL with 3 predominantly comprising BLs: DGG-BL (DDX3X, GNA13, and GNAI2), IC-BL (ID3 and CCND3), and Q53-BL (quiet TP53). Each BL subgroup is characterized by combinations of common driver and noncoding mutations caused by aberrant somatic hypermutation. The largest subgroups of BL cases, IC-BL and DGG-BL, are further characterized by distinct biological and gene expression differences. IC-BL and DGG-BL and their prototypical genetic features (ID3 and TP53) had significant associations with patient outcomes that were different among aBL and pBL cohorts. These findings highlight shared pathogenesis between aBL and pBL, and establish genetic subtypes within BL that serve to delineate tumors with distinct molecular features, providing a new framework for epidemiologic, diagnostic, and therapeutic strategies.
Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: R.D.M. and D.W.S. are named inventors on a patent application describing the double-hit signature. C.G.M. received research funding from Pfizer and AbbVie; was an advisory board member at Illumina; and was on the speaker’s bureau at Amgen. R.Y. reports receiving research support from Celgene (now Bristol Myers Squibb) through CRADAs with the NCI. R.Y. also reports receiving drugs for clinical trials from Merck, EMD-Serano, Eli Lilly, and CTI BioPharma through CRADAs with the NCI, and he has received drug supply for laboratory research from Janssen Pharmaceuticals. R.Y. is a coinventor on US Patent 10 001 483 entitled “Methods for the treatment of Kaposi's sarcoma or KSHV-induced lymphoma using immunomodulatory compounds and uses of biomarkers.” An immediate family member of R.Y. is a coinventor on patents or patent applications related to internalization of target receptors, epigenetic analysis, and ephrin tyrosine kinase inhibitors. All rights, title, and interest to these patents have been assigned to the US Department of Health and Human Services; the government conveys a portion of the royalties it receives to its employee inventors under the Federal Technology Transfer Act of 1986 (P.L. 99-502). A.N. received research funding from Pharmacyclics/AbbVie, Kite/Gilead, and Cornerstone; was a consultant for Janssen, Morphosys, Cornerstone, Epizyme, EUSA Pharma, TG Therapeutics, ADC Therapeutics, and Astra Zeneca; and has received honoraria from Pharmacyclics/AbbVie. The remaining authors declare no competing financial interests.
Figures


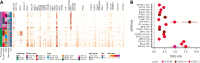
Comment in
-
Meet the Burkitts: a dark zone family.Blood. 2023 Feb 23;141(8):816-818. doi: 10.1182/blood.2022018509. Blood. 2023. PMID: 36821186 No abstract available.
References
Publication types
MeSH terms
Grants and funding
- HHSN261201100007I/CA/NCI NIH HHS/United States
- P01 CA019014/CA/NCI NIH HHS/United States
- 75N91019D00024/CA/NCI NIH HHS/United States
- R35 CA197695/CA/NCI NIH HHS/United States
- HHSN261201100063C/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- HHSN261201100007C/HL/NHLBI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- UM1 CA121947/CA/NCI NIH HHS/United States
- 75N91020C00003/CA/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

