Convergent and divergent biosynthetic strategies towards phosphonic acid natural products
- PMID: 36202046
- PMCID: PMC9722595
- DOI: 10.1016/j.cbpa.2022.102214
Convergent and divergent biosynthetic strategies towards phosphonic acid natural products
Abstract
The phosphonate class of natural products have received significant interests in the post-genomic era due to the relative ease with which their biosynthetic genes may be identified and the resultant final products be characterized. Recent large-scale studies of the elucidation and distributions of phosphonate pathways have provided a robust landscape for deciphering the underlying biosynthetic logic. A recurrent theme in phosphonate biosynthetic pathways is the interweaving of enzymatic reactions across different routes, which enables diversification to elaborate chemically novel scaffolds. Here, we provide a few vignettes of how Nature has utilized both convergent and divergent biosynthetic strategies to compile pathways for production of novel phosphonates. These examples illustrate how common intermediates may either be generated or intercepted to diversify chemical scaffolds and provides a starting point for both biotechnological and synthetic biological applications towards new phosphonates by similar combinatorial approaches.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


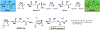
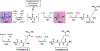

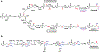
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

