Prevalence of Molecular Alterations in a Swiss Cohort of 512 Colorectal Carcinoma Patients by Targeted Next-Generation Sequencing Analysis in Routine Diagnostics
- PMID: 36202073
- PMCID: PMC10273900
- DOI: 10.1159/000526117
Prevalence of Molecular Alterations in a Swiss Cohort of 512 Colorectal Carcinoma Patients by Targeted Next-Generation Sequencing Analysis in Routine Diagnostics
Abstract
Introduction: Colorectal carcinoma (CRC) is among the most common carcinomas in women and men. In the advanced stage, patients are treated based on the RAS status. Recent studies indicate that in the future, in addition to KRAS and NRAS, alterations in other genes, such as PIK3CA or TP53, will be considered for therapy. Therefore, it is important to know the mutational landscape of routinely diagnosed CRC.
Method: We report the molecular profile of 512 Swiss CRC patients analyzed by targeted next-generation sequencing as part of routine diagnostics at our institute.
Results: Pathogenic and likely pathogenic variants were found in 462 (90%) CRC patients. Variants were detected in TP53 (54.3%), KRAS (48.2%), PIK3CA (15.6%), BRAF (13.5%), SMAD4 (10.5%), FBXW7 (7.8%), NRAS (3.5%), PTEN (2.7%), ERBB2 (1.6%), AKT1 (1.5%), and CTNNB1 (0.9%). The remaining pathogenic alterations were found in the genes ATM(n= 1), MAP2K1(n= 1), and IDH2(n= 1).
Discussion/conclusions: Our analysis revealed the prevalence of potential predictive markers in a large cohort of CRC patients obtained during routine diagnostic analysis. Furthermore, our study is the first of this size to uncover the molecular landscape of CRC in Switzerland.
Keywords: BRAF; Biomarkers; Cetuximab; Chromosome instability; Colon cancer; EGFR; KRAS; Microsatellite instability; Panitumumab.
© 2022 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
Matthias S. Matter has served as a consultant for Novartis and Glaxo Smith Cline and received speaker's honoraria from Thermo Fisher and Merck, all outside the current work. The other authors, namely, Simon Haefliger, Katharina Marston, Ilaria Alborelli, Philip M. Jermann, Edouard-Jean Stauffer, Mathias Gugger, Sylvia Hoeller, Luigi Tornillo, Luigi M. Terracciano, and Michel Bihl have no conflicts of interest to declare.
Figures


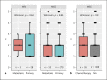
References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136((5)):E359–86. - PubMed
-
- Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27((8)):1386–1422. - PubMed
-
- Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19((3)):329–359. - PubMed
-
- You B, Chen EX. Anti-EGFR monoclonal antibodies for treatment of colorectal cancers: development of cetuximab and panitumumab. J Clin Pharmacol. 2012;52((2)):128–155. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

