Discovery, Chemistry, and Preclinical Development of Pritelivir, a Novel Treatment Option for Acyclovir-Resistant Herpes Simplex Virus Infections
- PMID: 36202389
- PMCID: PMC9620171
- DOI: 10.1021/acs.jmedchem.2c00668
Discovery, Chemistry, and Preclinical Development of Pritelivir, a Novel Treatment Option for Acyclovir-Resistant Herpes Simplex Virus Infections
Abstract
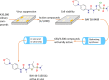
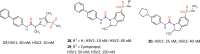
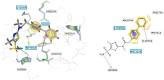
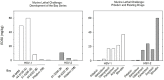
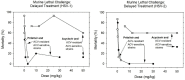
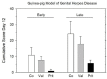
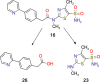
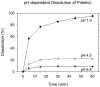
When the nucleoside analogue acyclovir was introduced in the early 1980s, it presented a game-changing treatment modality for herpes simplex virus infections. Since then, work has been ongoing to improve the weaknesses that have now been identified: a narrow time window for therapeutic success, resistance in immunocompromised patients, little influence on frequency of recurrences, relatively fast elimination, and poor bioavailability. The present Drug Annotation focuses on the helicase-primase inhibitor pritelivir currently in development for the treatment of acyclovir-resistant HSV infections and describes how a change of the molecular target (from viral DNA polymerase to the HSV helicase-primase complex) afforded improvement of the shortcomings of nucleoside analogs. Details are presented for the discovery process leading to the final drug candidate, the pivotal preclinical studies on mechanism of action and efficacy, and on how ongoing clinical research has been able to translate preclinical promises into clinical use.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors are or have been employees at AiCuris, the company that develops pritelivir.
Figures



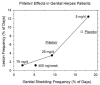
Similar articles
-
Helicase-primase as a target of new therapies for herpes simplex virus infections.Clin Pharmacol Ther. 2015 Jan;97(1):66-78. doi: 10.1002/cpt.3. Epub 2014 Nov 18. Clin Pharmacol Ther. 2015. PMID: 25670384 Review.
-
Efficacy of pritelivir and acyclovir in the treatment of herpes simplex virus infections in a mouse model of herpes simplex encephalitis.Antiviral Res. 2018 Jan;149:1-6. doi: 10.1016/j.antiviral.2017.11.002. Epub 2017 Nov 4. Antiviral Res. 2018. PMID: 29113740 Free PMC article.
-
Pharmacokinetics-pharmacodynamics of the helicase-primase inhibitor pritelivir following treatment of wild-type or pritelivir-resistant virus infection in a murine herpes simplex virus 1 infection model.Antimicrob Agents Chemother. 2014 Jul;58(7):3843-52. doi: 10.1128/AAC.02641-14. Epub 2014 Apr 21. Antimicrob Agents Chemother. 2014. PMID: 24752278 Free PMC article.
-
Oral bioavailability and in vivo efficacy of the helicase-primase inhibitor BILS 45 BS against acyclovir-resistant herpes simplex virus type 1.Antimicrob Agents Chemother. 2003 Jun;47(6):1798-804. doi: 10.1128/AAC.47.6.1798-1804.2003. Antimicrob Agents Chemother. 2003. PMID: 12760851 Free PMC article.
-
Antiviral agents for herpes simplex virus.Adv Pharmacol. 2013;67:1-38. doi: 10.1016/B978-0-12-405880-4.00001-9. Adv Pharmacol. 2013. PMID: 23885997 Review.
Cited by
-
In vitro evaluation of inhibitory effect of Lactobacillus reuteri supernatant on the replication of herpes simplex virus type 1 and expression of UL54, UL52 and UL27 genes.Iran J Microbiol. 2024 Feb;16(1):90-96. doi: 10.18502/ijm.v16i1.14877. Iran J Microbiol. 2024. PMID: 38682053 Free PMC article.
-
Optimizing Antiviral Dosing for HSV and CMV Treatment in Immunocompromised Patients.Pharmaceutics. 2023 Jan 3;15(1):163. doi: 10.3390/pharmaceutics15010163. Pharmaceutics. 2023. PMID: 36678792 Free PMC article. Review.
-
Five-Membered Heterocyclic Sulfonamides as Carbonic Anhydrase Inhibitors.Molecules. 2023 Apr 4;28(7):3220. doi: 10.3390/molecules28073220. Molecules. 2023. PMID: 37049983 Free PMC article. Review.
-
A Multicenter Assessment of the Outcomes and Toxicities of Foscarnet for Treatment of Acyclovir-Resistant Mucocutaneous Herpes Simplex in Immunocompromised Patients.Open Forum Infect Dis. 2024 Feb 5;11(3):ofae046. doi: 10.1093/ofid/ofae046. eCollection 2024 Mar. Open Forum Infect Dis. 2024. PMID: 38444818 Free PMC article.
-
Structural Chemistry of Helicase Inhibition.J Med Chem. 2025 Feb 27;68(4):4022-4039. doi: 10.1021/acs.jmedchem.4c01909. Epub 2025 Feb 11. J Med Chem. 2025. PMID: 39933052 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information
Medical

