Consequences of telomere dysfunction in fibroblasts, club and basal cells for lung fibrosis development
- PMID: 36202783
- PMCID: PMC9537293
- DOI: 10.1038/s41467-022-32771-6
Consequences of telomere dysfunction in fibroblasts, club and basal cells for lung fibrosis development
Abstract
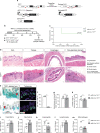
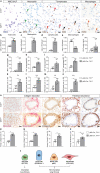
TRF1 is an essential component of the telomeric protective complex or shelterin. We previously showed that dysfunctional telomeres in alveolar type II (ATII) cells lead to interstitial lung fibrosis. Here, we study the lung pathologies upon telomere dysfunction in fibroblasts, club and basal cells. TRF1 deficiency in lung fibroblasts, club and basal cells induced telomeric damage, proliferative defects, cell cycle arrest and apoptosis. While Trf1 deletion in fibroblasts does not spontaneously lead to lung pathologies, upon bleomycin challenge exacerbates lung fibrosis. Unlike in females, Trf1 deletion in club and basal cells from male mice resulted in lung inflammation and airway remodeling. Here, we show that depletion of TRF1 in fibroblasts, Club and basal cells does not lead to interstitial lung fibrosis, underscoring ATII cells as the relevant cell type for the origin of interstitial fibrosis. Our findings contribute to a better understanding of proper telomere protection in lung tissue homeostasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Renal fibroblasts are involved in fibrogenic changes in kidney fibrosis associated with dysfunctional telomeres.Exp Mol Med. 2024 Oct;56(10):2216-2230. doi: 10.1038/s12276-024-01318-8. Epub 2024 Oct 1. Exp Mol Med. 2024. PMID: 39349834 Free PMC article.
-
Telomere dysfunction in alveolar epithelial cells causes lung remodeling and fibrosis.JCI Insight. 2016 Sep 8;1(14):e86704. doi: 10.1172/jci.insight.86704. JCI Insight. 2016. PMID: 27699234 Free PMC article.
-
Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice.Genes Dev. 2009 Sep 1;23(17):2060-75. doi: 10.1101/gad.543509. Epub 2009 Aug 13. Genes Dev. 2009. PMID: 19679647 Free PMC article.
-
Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance.Mech Ageing Dev. 2012 Jun;133(6):421-34. doi: 10.1016/j.mad.2012.05.002. Epub 2012 May 23. Mech Ageing Dev. 2012. PMID: 22634377 Review.
-
Club Cells-A Guardian against Occupational Hazards.Biomedicines. 2023 Dec 28;12(1):78. doi: 10.3390/biomedicines12010078. Biomedicines. 2023. PMID: 38255185 Free PMC article. Review.
Cited by
-
Senescence of alveolar epithelial progenitor cells: a critical driver of lung fibrosis.Am J Physiol Cell Physiol. 2023 Aug 1;325(2):C483-C495. doi: 10.1152/ajpcell.00239.2023. Epub 2023 Jul 17. Am J Physiol Cell Physiol. 2023. PMID: 37458437 Free PMC article. Review.
-
Short airway telomeres are associated with primary graft dysfunction and chronic lung allograft dysfunction.J Heart Lung Transplant. 2023 Dec;42(12):1700-1709. doi: 10.1016/j.healun.2023.08.018. Epub 2023 Aug 28. J Heart Lung Transplant. 2023. PMID: 37648073 Free PMC article.
-
Different Role of TRF1 and TRF2 Expression in Non-Small Cell Lung Cancers.Onco Targets Ther. 2024 Jun 4;17:463-469. doi: 10.2147/OTT.S461430. eCollection 2024. Onco Targets Ther. 2024. PMID: 38855632 Free PMC article.
-
Regenerative Capacity of Alveolar Type 2 Cells Is Proportionally Reduced Following Disease Progression in Idiopathic Pulmonary Fibrosis-Derived Organoid Cultures.Tuberc Respir Dis (Seoul). 2025 Jan;88(1):130-137. doi: 10.4046/trd.2024.0094. Epub 2024 Sep 27. Tuberc Respir Dis (Seoul). 2025. PMID: 39343426 Free PMC article.
-
A new variant in the ZCCHC8 gene: diverse clinical phenotypes and expression in the lung.ERJ Open Res. 2024 Feb 19;10(1):00487-2023. doi: 10.1183/23120541.00487-2023. eCollection 2024 Jan. ERJ Open Res. 2024. PMID: 38375433 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

