DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2
- PMID: 36202799
- PMCID: PMC9537531
- DOI: 10.1038/s41467-022-33309-6
DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2
Abstract
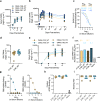
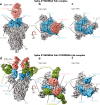
Monoclonal antibody therapy has played an important role against SARS-CoV-2. Strategies to deliver functional, antibody-based therapeutics with improved in vivo durability are needed to supplement current efforts and reach underserved populations. Here, we compare recombinant mAbs COV2-2196 and COV2-2130, which compromise clinical cocktail Tixagevimab/Cilgavimab, with optimized nucleic acid-launched forms. Functional profiling of in vivo-expressed, DNA-encoded monoclonal antibodies (DMAbs) demonstrated similar specificity, broad antiviral potency and equivalent protective efficacy in multiple animal challenge models of SARS-CoV-2 prophylaxis compared to protein delivery. In PK studies, DNA-delivery drove significant serum antibody titers that were better maintained compared to protein administration. Furthermore, cryo-EM studies performed on serum-derived DMAbs provide the first high-resolution visualization of in vivo-launched antibodies, revealing new interactions that may promote cooperative binding to trimeric antigen and broad activity against VoC including Omicron lineages. These data support the further study of DMAb technology in the development and delivery of valuable biologics.
© 2022. The Author(s).
Conflict of interest statement
K.S., T.R.F.S., V.M.A., I.M., K.E.B. and J.D.B. are employees of Inovio Pharmaceuticals and as such receives salary and benefits, including ownership of stock and stock options, from the company. J.F, K.R, K.R and M.T.E are employees of and hold or may hold stock in AstraZeneca. S.K.W. is an inventor on US patent (US20190216949) for the AAV6.2FF capsid. D.B.W. has received grant funding, participates in industry collaborations, has received speaking honoraria, and has received fees for consulting, including serving on scientific review committees and board series. Remuneration received by D.B.W. includes direct payments and stock or stock options. D.B.W. also discloses the following paid associations with commercial partners: GeneOne (consultant), Geneos (advisory board), AstraZeneca (advisory board, speaker), Inovio (BOD, SRA, Stock), Sanofi (advisory board) and BBI (advisory board). All other authors declare no completing interests.
Figures





References
-
- Johns Hopkins Coronavirus Resource Center. COVID-19 Map. https://coronavirus.jhu.edu/map.html. (2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

