RNA export through the nuclear pore complex is directional
- PMID: 36202822
- PMCID: PMC9537521
- DOI: 10.1038/s41467-022-33572-7
RNA export through the nuclear pore complex is directional
Abstract
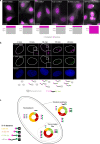
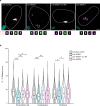
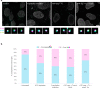
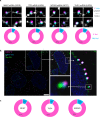
The changes occurring in mRNA organization during nucleo-cytoplasmic transport and export, are not well understood. Moreover, directionality of mRNA passage through the nuclear pore complex (NPC) has not been examined within individual NPCs. Here we find that an mRNP is compact during nucleoplasmic travels compared to a more open structure after transcription and at the nuclear periphery. Compaction levels of nuclear transcripts can be modulated by varying levels of SR proteins and by changing genome organization. Nuclear mRNPs are mostly rod-shaped with distant 5'/3'-ends, although for some, the ends are in proximity. The latter is more abundant in the cytoplasm and can be modified by translation inhibition. mRNAs and lncRNAs exiting the NPC exhibit predominant 5'-first export. In some cases, several adjacent NPCs are engaged in export of the same mRNA suggesting 'gene gating'. Altogether, we show that the mRNP is a flexible structure during travels, with 5'-directionality during export.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

