Deep Learning-based Assessment of Oncologic Outcomes from Natural Language Processing of Structured Radiology Reports
- PMID: 36204531
- PMCID: PMC9530771
- DOI: 10.1148/ryai.220055
Deep Learning-based Assessment of Oncologic Outcomes from Natural Language Processing of Structured Radiology Reports
Abstract
Purpose: To train a deep natural language processing (NLP) model, using data mined structured oncology reports (SOR), for rapid tumor response category (TRC) classification from free-text oncology reports (FTOR) and to compare its performance with human readers and conventional NLP algorithms.
Materials and methods: In this retrospective study, databases of three independent radiology departments were queried for SOR and FTOR dated from March 2018 to August 2021. An automated data mining and curation pipeline was developed to extract Response Evaluation Criteria in Solid Tumors-related TRCs for SOR for ground truth definition. The deep NLP bidirectional encoder representations from transformers (BERT) model and three feature-rich algorithms were trained on SOR to predict TRCs in FTOR. Models' F1 scores were compared against scores of radiologists, medical students, and radiology technologist students. Lexical and semantic analyses were conducted to investigate human and model performance on FTOR.
Results: Oncologic findings and TRCs were accurately mined from 9653 of 12 833 (75.2%) queried SOR, yielding oncology reports from 10 455 patients (mean age, 60 years ± 14 [SD]; 5303 women) who met inclusion criteria. On 802 FTOR in the test set, BERT achieved better TRC classification results (F1, 0.70; 95% CI: 0.68, 0.73) than the best-performing reference linear support vector classifier (F1, 0.63; 95% CI: 0.61, 0.66) and technologist students (F1, 0.65; 95% CI: 0.63, 0.67), had similar performance to medical students (F1, 0.73; 95% CI: 0.72, 0.75), but was inferior to radiologists (F1, 0.79; 95% CI: 0.78, 0.81). Lexical complexity and semantic ambiguities in FTOR influenced human and model performance, revealing maximum F1 score drops of -0.17 and -0.19, respectively.
Conclusion: The developed deep NLP model reached the performance level of medical students but not radiologists in curating oncologic outcomes from radiology FTOR.Keywords: Neural Networks, Computer Applications-Detection/Diagnosis, Oncology, Research Design, Staging, Tumor Response, Comparative Studies, Decision Analysis, Experimental Investigations, Observer Performance, Outcomes Analysis Supplemental material is available for this article. © RSNA, 2022.
Keywords: Comparative Studies; Computer Applications–Detection/Diagnosis; Decision Analysis; Experimental Investigations; Neural Networks; Observer Performance; Oncology; Outcomes Analysis; Research Design; Staging; Tumor Response.
© 2022 by the Radiological Society of North America, Inc.
Conflict of interest statement
Disclosures of conflicts of interest: M.A.F. No relevant relationships. K.K. No relevant relationships. A.B. No relevant relationships. M.M. No relevant relationships. M.S. No relevant relationships. M.K. No relevant relationships. G.K. No relevant relationships. J.S. No relevant relationships. C.P.H. No relevant relationships. H.P.S. No relevant relationships. K.M.H. No relevant relationships. T.F.W. No relevant relationships. J.K. No relevant relationships.
Figures

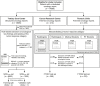
![Structured oncologic assessment in clinical routine and natural language processing (NLP) model building. An exemplary structured oncology report (SOR) for a 32-year-old woman with a history of breast cancer (left side) was interpreted as progressive disease (PD). The oncologic data were automatically processed and then fed into the NLP development pipeline (right side, A–E). (A) The deep NLP architecture used was based on the bidirectional encoder representations from transformers (BERT) language model pretrained on unlabeled general domain data and adapted to the German vocabulary. (B) Automatic extraction of the Response Evaluation Criteria in Solid Tumors (RECIST)–related categories PD, stable disease (SD), partial response (PR), and complete response (CR) from the SOR “impression” section by using a rule-based pattern-matching command called regular expressions (RegEx). (C) Fine-tuning of BERT and three feature-rich NLP methods (linear support vector classifier [SVC], k-nearest neighbors [KNN], multinomial naive Bayes [MNB]) on the extracted SOR oncologic findings section. The output of (B) was used as ground truth classifier for (D) NLP model training and validation, followed by (E) performance evaluation on the free-text oncology reports (FTOR) test sets in comparison with human baseline scores. A live demo of the SOR template can be accessed for review at http://www.targetedreporting.com/sor/. For demonstration purposes, the presented exemplary SOR and the online template have been translated from German to English. TF-IDF = term frequency–inverse document frequency.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e688/9530771/7b9968825374/ryai.220055.fig2.gif)



References
-
- Yim WW , Yetisgen M , Harris WP , Kwan SW . Natural language processing in oncology: a review . JAMA Oncol 2016. ; 2 ( 6 ): 797 – 804 . - PubMed
-
- Fink MA , Mayer VL , Schneider T , et al. . CT angiography clot burden score from data mining of structured reports for pulmonary embolism . Radiology 2022. ; 302 ( 1 ): 175 – 184 . - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

