Isolation of Pseudomonas aromaticivorans sp. nov from a hydrocarbon-contaminated groundwater capable of degrading benzene-, toluene-, m- and p-xylene under microaerobic conditions
- PMID: 36204622
- PMCID: PMC9530055
- DOI: 10.3389/fmicb.2022.929128
Isolation of Pseudomonas aromaticivorans sp. nov from a hydrocarbon-contaminated groundwater capable of degrading benzene-, toluene-, m- and p-xylene under microaerobic conditions
Abstract
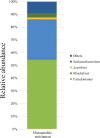
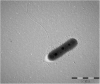
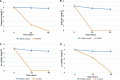
Members of the genus Pseudomonas are known to be widespread in hydrocarbon contaminated environments because of their remarkable ability to degrade a variety of petroleum hydrocarbons, including BTEX (benzene, toluene, ethylbenzene and xylene) compounds. During an enrichment investigation which aimed to study microaerobic xylene degradation in a legacy petroleum hydrocarbon-contaminated groundwater, a novel Gram-stain-negative, aerobic, motile and rod-shaped bacterial strain, designated as MAP12T was isolated. It was capable of degrading benzene, toluene, meta- and para- xylene effectively under both aerobic and microaerobic conditions. The 16S rRNA gene sequence analysis revealed that strain MAP12T belongs to the genus Pseudomonas, with the highest 16S rRNA gene similarity to Pseudomonas linyingensis LYBRD3-7 T (98.42%), followed by Pseudomonas sagittaria JCM 18195 T (98.29%) and Pseudomonas alcaliphila JCM 10630 T (98.08%). Phylogenomic tree constructed using a concatenated alignment of 92 core genes indicated that strain MAP12T is distinct from any known Pseudomonas species. The draft genome sequence of strain MAP12T is 4.36 Mb long, and the G+C content of MAP12T genome is 65.8%. Orthologous average nucleotide identity (OrthoANI) and digital DNA-DNA hybridization (dDDH) analyses confirmed that strain MAP12T is distinctly separated from its closest neighbors (OrthoANI < 89 %; dDDH < 36%). Though several members of the genus Pseudomonas are well known for their aerobic BTEX degradation capability, this is the first report of a novel Pseudomonas species capable of degrading xylene under microaerobic conditions. By applying genome-resolved metagenomics, we were able to partially reconstruct the genome of strain MAP12 T from metagenomics sequence data and showed that strain MAP12 T was an abundant member of the xylene-degrading bacterial community under microaerobic conditions. Strain MAP12T contains ubiquinone 9 (Q9) as the major respiratory quinone and diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine as major polar lipids. The major cellular fatty acids of strain MAP12T are summed feature 3 (C16:1ω6c and/or C16:1ω7c), C16:0 and summed feature 8 (C18:1ω6c and/or C18:1ω7c). The results of this polyphasic study support that strain MAP12T represents a novel species of the genus Pseudomonas, hence the name of Pseudomonas aromaticivorans sp. nov. is proposed for this strain considering its aromatic hydrocarbon degradation capability. The type strain is MAP12T (=LMG 32466, =NCAIM B.02668).
Keywords: BTEX; Pseudomona; biodegradation; groundwater; xylene.
Copyright © 2022 Banerjee, Bedics, Tóth, Kriszt, Soares, Bóka and Táncsics.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Banerjee S., Bedics A., Harkai P., Kriszt B., Alpula N., Táncsics A. (2022). Evaluating the aerobic xylene-degrading potential of the intrinsic microbial community of a legacy BTEX-contaminated aquifer by enrichment culturing coupled with multi-omics analysis: uncovering the role of Hydrogenophaga strains in xylene degradation. Environ. Sci. Pollut. Res. 29 28431–28445. 10.1007/s11356-021-18300-w - DOI - PMC - PubMed
-
- Banerjee S., Táncsics A., Tóth E., Révész F., Bóka K., Kriszt B. (2021). Hydrogenophaga aromaticivorans sp. nov., isolated from a para-xylene-degrading enrichment culture, capable of degrading benzene, meta-and para-xylene. Int. J. Syst. Evol. Microbiol. 71:004743. 10.1099/ijsem.0.004743 - DOI - PubMed
-
- Barrow G. I., Feltham R. K. A. (1993). Cowan and Steel’s Manual for the Identification of Medical Bacteria, 3rd Edn. Cambridge: Cambridge University Press, 10.1017/CBO9780511527104 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

