Mining Potential Drug Targets and Constructing Diagnostic Models for Heart Failure Based on miRNA-mRNA Networks
- PMID: 36204659
- PMCID: PMC9532133
- DOI: 10.1155/2022/9652169
Mining Potential Drug Targets and Constructing Diagnostic Models for Heart Failure Based on miRNA-mRNA Networks
Abstract
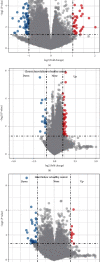
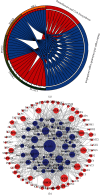
Heart failure (HF) is a globally prevalent cardiovascular disease, but effective drug targets and diagnostic models are still lacking. This study was designed to investigate effective drug targets and diagnostic models for HF in terms of miRNA targets, hoping to contribute to the understanding and treatment of HF. Using HF miRNA and gene expression profile data from the GEO database, we analyzed differentially expressed miRNAs/gene identification in HF using Limma and predicted miRNA targets by the online TargetScan database. Subsequently, gene set enrichment analysis and annotation were performed using WebGestaltR package. Protein-protein interactions were identified using the STRING database. The proximity of drugs to treat HF was also calculated and predicted for potential target therapeutic drug. In addition, further drug identification was performed by molecular docking. Finally, diagnostic models were constructed based on differential miRNAs. The GEO dataset was used to screen 66 differentially expressed miRNAs, incorporating 56 downregulated miRNAs and 10 upregulated miRNAs. The JAK-STAT signaling pathway, MAPK signaling pathway, p53 signaling pathway, Prolactin signaling pathway, and TGF-beta signaling pathway were enriched, as shown by KEGG enrichment analysis on the target genes. In addition, we found that 83 genes were upregulated and 92 genes were downregulated in HF patients vs. healthy individuals. Based on the inflammation-related score, hypoxia-related score, and energy metabolism-related score, we identified key miRNA-mRNA pairs and constructed an interaction network. Following that, TAP1, which had the highest expression and network connectivity in acute HF with crystal and molecular docking studies, was selected as a key candidate gene in the network. And the compound DB04847 was selected to produce a large number of favorable interactions with TAP1 protein. Finally, we constructed two diagnostic models based on the differential miRNAs hsa-miR-6785-5p and hsa-miR-4443. In conclusion, we identified TAP1, a key candidate gene in the diagnosis and treatment of HF, and determined that compound DB04847 is highly likely to be a potential inhibitor of TAP1. The TAP1 gene was also found to be regulated by hsa-miR-6785-5p and hsa-miR-4443, and a diagnostic model was constructed. This provides a new promising direction to improve the diagnosis, prognosis, and treatment outcome and guide more effective immunotherapy strategies of HF.
Copyright © 2022 Xiangming Fang et al.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures






References
-
- Shah S. J., Feldman T., Ricciardi M. J., et al. One-year safety and clinical outcomes of a transcatheter interatrial shunt device for the treatment of heart failure with preserved ejection fraction in the reduce elevated left atrial pressure in patients with heart failure (REDUCE LAP-HF I) trial. JAMA Cardiology . 2018;3(10):968–977. doi: 10.1001/jamacardio.2018.2936. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

