Unbiased proteomic and forward genetic screens reveal that mechanosensitive ion channel MSL10 functions at ER-plasma membrane contact sites in Arabidopsis thaliana
- PMID: 36205399
- PMCID: PMC9625087
- DOI: 10.7554/eLife.80501
Unbiased proteomic and forward genetic screens reveal that mechanosensitive ion channel MSL10 functions at ER-plasma membrane contact sites in Arabidopsis thaliana
Abstract
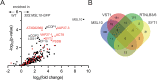
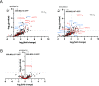
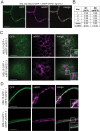
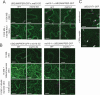
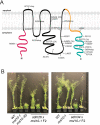
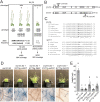
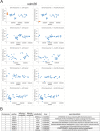
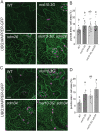
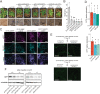
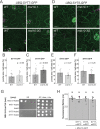
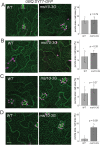
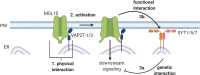
Mechanosensitive (MS) ion channels are an evolutionarily conserved way for cells to sense mechanical forces and transduce them into ionic signals. The channel properties of Arabidopsis thaliana MscS-Like (MSL)10 have been well studied, but how MSL10 signals remains largely unknown. To uncover signaling partners of MSL10, we employed a proteomic screen and a forward genetic screen; both unexpectedly implicated endoplasmic reticulum-plasma membrane contact sites (EPCSs) in MSL10 function. The proteomic screen revealed that MSL10 associates with multiple proteins associated with EPCSs. Of these, only VAMP-associated proteins (VAP)27-1 and VAP27-3 interacted directly with MSL10. The forward genetic screen, for suppressors of a gain-of-function MSL10 allele (msl10-3G, MSL10S640L), identified mutations in the synaptotagmin (SYT)5 and SYT7 genes. We also found that EPCSs were expanded in leaves of msl10-3G plants compared to the wild type. Taken together, these results indicate that MSL10 associates and functions with EPCS proteins, providing a new cell-level framework for understanding MSL10 signaling. In addition, placing a mechanosensory protein at EPCSs provides new insight into the function and regulation of this type of subcellular compartment.
Keywords: A. thaliana; VAMP-associated proteins; mechanosensitive ion channel; mechanotransduction; membrane contact sites; plant biology; synaptotagmins.
© 2022, Codjoe et al.
Conflict of interest statement
JC, RR, FM, RV, EH No competing interests declared
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

