Glycan Modulation of Insulin-like Growth Factor-1 Receptor
- PMID: 36205924
- PMCID: PMC9712175
- DOI: 10.1002/anie.202211320
Glycan Modulation of Insulin-like Growth Factor-1 Receptor
Abstract
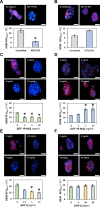
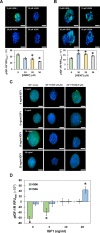
The insulin-like growth factor-1 receptor (IGF-1R) is a receptor tyrosine kinase (RTK) that plays critical roles in cancer. Microarray, computational, thermodynamic, and cellular imaging studies reveal that activation of IGF-1R by its cognate ligand IGF1 is inhibited by shorter, soluble heparan sulfate (HS) sequences (e.g., HS06), whereas longer polymeric chains do not inhibit the RTK, a phenomenon directly opposed to the traditional relationship known for GAG-protein systems. The inhibition arises from smaller oligosaccharides binding in a unique pocket in the IGF-1R ectodomain, which competes with the natural cognate ligand IGF1. This work presents a highly interesting observation on preferential and competing inhibition of IGF-1R by smaller sequences, whereas polysaccharides are devoid of this function. These insights will be of major value to glycobiologists and anti-cancer drug discoverers.
Keywords: Drug Discovery; Glycosaminoglycans; Growth Factors; Heparan Sulfate; Receptor Tyrosine Kinases.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

