A human iPSC-array-based GWAS identifies a virus susceptibility locus in the NDUFA4 gene and functional variants
- PMID: 36206731
- PMCID: PMC9550219
- DOI: 10.1016/j.stem.2022.09.008
A human iPSC-array-based GWAS identifies a virus susceptibility locus in the NDUFA4 gene and functional variants
Abstract
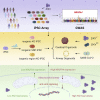
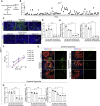
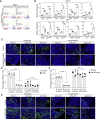
Population-based studies to identify disease-associated risk alleles typically require samples from a large number of individuals. Here, we report a human-induced pluripotent stem cell (hiPSC)-based screening strategy to link human genetics with viral infectivity. A genome-wide association study (GWAS) identified a cluster of single-nucleotide polymorphisms (SNPs) in a cis-regulatory region of the NDUFA4 gene, which was associated with susceptibility to Zika virus (ZIKV) infection. Loss of NDUFA4 led to decreased sensitivity to ZIKV, dengue virus, and SARS-CoV-2 infection. Isogenic hiPSC lines carrying non-risk alleles of SNPs or deletion of the cis-regulatory region lower sensitivity to viral infection. Mechanistic studies indicated that loss/reduction of NDUFA4 causes mitochondrial stress, which leads to the leakage of mtDNA and thereby upregulation of type I interferon signaling. This study provides proof-of-principle for the application of iPSC arrays in GWAS and identifies NDUFA4 as a previously unknown susceptibility locus for viral infection.
Keywords: Dengue Virus; NDUFA4; SARS-CoV-2; genome-wide association study; iPSC array; isogenic hiPSC lines; mtDNA; risk allele; single-nucleotide polymorphism; type I interferon.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.E.S. is on the scientific advisory board of Miromatrix, Inc and Lime Therapeutics and is a paid consultant and speaker for Alnylam, Inc. S.C. and T.E. are the co-founders of OncoBeat, LLC. S.C. is a consultant of Vesaliustx Therapeutics.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

