Technology meets TILs: Deciphering T cell function in the -omics era
- PMID: 36206755
- PMCID: PMC9839604
- DOI: 10.1016/j.ccell.2022.09.011
Technology meets TILs: Deciphering T cell function in the -omics era
Abstract
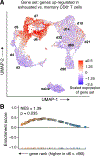
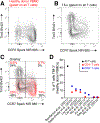
T cells are at the center of cancer immunology because of their ability to recognize mutations in tumor cells and directly mediate cancer cell killing. Immunotherapies to rejuvenate exhausted T cell responses have transformed the clinical management of several malignancies. In parallel, the development of novel multidimensional analysis platforms, such as single-cell RNA sequencing and high-dimensional flow cytometry, has yielded unprecedented insights into immune cell biology. This convergence has revealed substantial heterogeneity of tumor-infiltrating immune cells in single tumors, across tumor types, and among individuals with cancer. Here we discuss the opportunities and challenges of studying the complex tumor microenvironment with -omics technologies that generate vast amounts of data, highlighting the opportunities and limitations of these technologies with a particular focus on interpreting high-dimensional studies of CD8+ T cells in the tumor microenvironment.
Keywords: CD4; CD8; PD-1; T cell; bystander; cancer immunology; flow cytometry; immunotherapy; scRNA-seq; techniques; tumor-infiltrating lymphocytes.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Abdel-Hakeem MS, Manne S, Beltra J-C, Stelekati E, Chen Z, Nzingha K, Ali M-A, Johnson JL, Giles JR, Mathew D, et al. (2021). Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nature Immunology 22, 1008–1019. 10.1038/s41590-021-00975-5. - DOI - PMC - PubMed
-
- Acharya N, Madi A, Zhang H, Klapholz M, Escobar G, Dulberg S, Christian E, Ferreira M, Dixon KO, Fell G, et al. (2020). Endogenous Glucocorticoid Signaling Regulates CD8+ T Cell Differentiation and Development of Dysfunction in the Tumor Microenvironment. Immunity 53, 658–671.e656. 10.1016/j.immuni.2020.08.005. - DOI - PMC - PubMed
-
- Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, et al. (2009). The Yellow Fever Virus Vaccine Induces a Broad and Polyfunctional Human Memory CD8+ T Cell Response. The Journal of Immunology 183, 7919–7930. 10.4049/jimmunol.0803903. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

