Prevalence and mechanisms of somatic deletions in single human neurons during normal aging and in DNA repair disorders
- PMID: 36207339
- PMCID: PMC9546902
- DOI: 10.1038/s41467-022-33642-w
Prevalence and mechanisms of somatic deletions in single human neurons during normal aging and in DNA repair disorders
Abstract
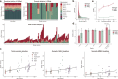
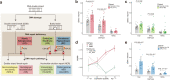
Replication errors and various genotoxins cause DNA double-strand breaks (DSBs) where error-prone repair creates genomic mutations, most frequently focal deletions, and defective repair may lead to neurodegeneration. Despite its pathophysiological importance, the extent to which faulty DSB repair alters the genome, and the mechanisms by which mutations arise, have not been systematically examined reflecting ineffective methods. Here, we develop PhaseDel, a computational method to detect focal deletions and characterize underlying mechanisms in single-cell whole genome sequences (scWGS). We analyzed high-coverage scWGS of 107 single neurons from 18 neurotypical individuals of various ages, and found that somatic deletions increased with age and in highly expressed genes in human brain. Our analysis of 50 single neurons from DNA repair-deficient diseases with progressive neurodegeneration (Cockayne syndrome, Xeroderma pigmentosum, and Ataxia telangiectasia) reveals elevated somatic deletions compared to age-matched controls. Distinctive mechanistic signatures and transcriptional associations suggest roles for somatic deletions in neurodegeneration.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

