Common anti-cancer therapies induce somatic mutations in stem cells of healthy tissue
- PMID: 36207433
- PMCID: PMC9546852
- DOI: 10.1038/s41467-022-33663-5
Common anti-cancer therapies induce somatic mutations in stem cells of healthy tissue
Abstract
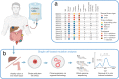
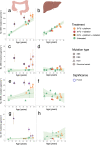
Genome-wide mutation analyses have revealed that specific anti-cancer drugs are highly mutagenic to cancer cells, but the mutational impact of anti-cancer therapies on normal cells is not known. Here, we examine genome-wide somatic mutation patterns in 42 healthy adult stem cells (ASCs) of the colon or the liver from 14 cancer patients (mean of 3.2 ASC per donor) that received systemic chemotherapy and/or local radiotherapy. The platinum-based chemo-drug Oxaliplatin induces on average 535 ± 260 mutations in colon ASC, while 5-FU shows a complete mutagenic absence in most, but not all colon ASCs. In contrast with the colon, normal liver ASCs escape mutagenesis from systemic treatment with Oxaliplatin and 5-FU. Thus, while chemotherapies are highly effective at killing cancer cells, their systemic use also increases the mutational burden of long-lived normal stem cells responsible for tissue renewal thereby increasing the risk for developing second cancers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

