MYC oncogene elicits tumorigenesis associated with embryonic, ribosomal biogenesis, and tissue-lineage dedifferentiation gene expression changes
- PMID: 36207533
- PMCID: PMC10257951
- DOI: 10.1038/s41388-022-02458-9
MYC oncogene elicits tumorigenesis associated with embryonic, ribosomal biogenesis, and tissue-lineage dedifferentiation gene expression changes
Abstract
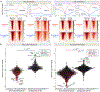
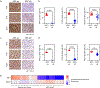
MYC is a transcription factor frequently overexpressed in cancer. To determine how MYC drives the neoplastic phenotype, we performed transcriptomic analysis using a panel of MYC-driven autochthonous transgenic mouse models. We found that MYC elicited gene expression changes mostly in a tissue- and lineage-specific manner across B-cell lymphoma, T-cell acute lymphoblastic lymphoma, hepatocellular carcinoma, renal cell carcinoma, and lung adenocarcinoma. However, despite these gene expression changes being mostly tissue-specific, we uncovered a convergence on a common pattern of upregulation of embryonic stem cell gene programs and downregulation of tissue-of-origin gene programs across MYC-driven cancers. These changes are representative of lineage dedifferentiation, that may be facilitated by epigenetic alterations that occur during tumorigenesis. Moreover, while several cellular processes are represented among embryonic stem cell genes, ribosome biogenesis is most specifically associated with MYC expression in human primary cancers. Altogether, MYC's capability to drive tumorigenesis in diverse tissue types appears to be related to its ability to both drive a core signature of embryonic genes that includes ribosomal biogenesis genes as well as promote tissue and lineage specific dedifferentiation.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Declaration of Interests
The authors have no conflicts of interest to disclose.
Figures




References
-
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002. Jul 5;297(5578):63–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

