SVD-CLAHE boosting and balanced loss function for Covid-19 detection from an imbalanced Chest X-Ray dataset
- PMID: 36208598
- PMCID: PMC9514969
- DOI: 10.1016/j.compbiomed.2022.106092
SVD-CLAHE boosting and balanced loss function for Covid-19 detection from an imbalanced Chest X-Ray dataset
Abstract
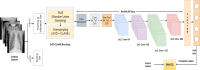
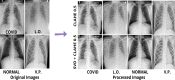
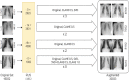
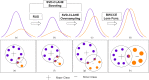
Covid-19 disease has had a disastrous effect on the health of the global population, for the last two years. Automatic early detection of Covid-19 disease from Chest X-Ray (CXR) images is a very crucial step for human survival against Covid-19. In this paper, we propose a novel data-augmentation technique, called SVD-CLAHE Boosting and a novel loss function Balanced Weighted Categorical Cross Entropy (BWCCE), in order to detect Covid 19 disease efficiently from a highly class-imbalanced Chest X-Ray image dataset. Our proposed SVD-CLAHE Boosting method is comprised of both oversampling and under-sampling methods. First, a novel Singular Value Decomposition (SVD) based contrast enhancement and Contrast Limited Adaptive Histogram Equalization (CLAHE) methods are employed for oversampling the data in minor classes. Simultaneously, a Random Under Sampling (RUS) method is incorporated in major classes, so that the number of images per class will be more balanced. Thereafter, Balanced Weighted Categorical Cross Entropy (BWCCE) loss function is proposed in order to further reduce small class imbalance after SVD-CLAHE Boosting. Experimental results reveal that ResNet-50 model on the augmented dataset (by SVD-CLAHE Boosting), along with BWCCE loss function, achieved 95% F1 score, 94% accuracy, 95% recall, 96% precision and 96% AUC, which is far better than the results by other conventional Convolutional Neural Network (CNN) models like InceptionV3, DenseNet-121, Xception etc. as well as other existing models like Covid-Lite and Covid-Net. Hence, our proposed framework outperforms other existing methods for Covid-19 detection. Furthermore, the same experiment is conducted on VGG-19 model in order to check the validity of our proposed framework. Both ResNet-50 and VGG-19 model are pre-trained on the ImageNet dataset. We publicly shared our proposed augmented dataset on Kaggle website (https://www.kaggle.com/tr1gg3rtrash/balanced-augmented-covid-cxr-dataset), so that any research community can widely utilize this dataset. Our code is available on GitHub website online (https://github.com/MrinalTyagi/SVD-CLAHE-and-BWCCE).
Keywords: Categorical Cross Entropy (CCE); Chest X-Ray (CXR) images; Class imbalance problem; Contrast Limited Adaptive Histogram Equalization (CLAHE); Covid-19 detection; Data augmentation; Singular Value Decomposition (SVD).
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images.Comput Biol Med. 2021 May;132:104319. doi: 10.1016/j.compbiomed.2021.104319. Epub 2021 Mar 11. Comput Biol Med. 2021. PMID: 33799220 Free PMC article.
-
LCSB-inception: Reliable and effective light-chroma separated branches for Covid-19 detection from chest X-ray images.Comput Biol Med. 2022 Nov;150:106195. doi: 10.1016/j.compbiomed.2022.106195. Epub 2022 Oct 14. Comput Biol Med. 2022. PMID: 37859288 Free PMC article.
-
COVID-19 detection on chest X-ray images using Homomorphic Transformation and VGG inspired deep convolutional neural network.Biocybern Biomed Eng. 2023 Jan-Mar;43(1):1-16. doi: 10.1016/j.bbe.2022.11.003. Epub 2022 Nov 24. Biocybern Biomed Eng. 2023. PMID: 36447948 Free PMC article.
-
CLAHE-CapsNet: Efficient retina optical coherence tomography classification using capsule networks with contrast limited adaptive histogram equalization.PLoS One. 2023 Nov 30;18(11):e0288663. doi: 10.1371/journal.pone.0288663. eCollection 2023. PLoS One. 2023. PMID: 38032915 Free PMC article. Review.
-
Development and integration of VGG and dense transfer-learning systems supported with diverse lung images for discovery of the Coronavirus identity.Inform Med Unlocked. 2022;32:101004. doi: 10.1016/j.imu.2022.101004. Epub 2022 Jul 8. Inform Med Unlocked. 2022. PMID: 35822170 Free PMC article. Review.
Cited by
-
Automated classification of chest X-rays: a deep learning approach with attention mechanisms.BMC Med Imaging. 2025 Mar 4;25(1):71. doi: 10.1186/s12880-025-01604-5. BMC Med Imaging. 2025. PMID: 40038588 Free PMC article.
-
Improving Image Quality of Chest Radiography with Artificial Intelligence-Supported Dual-Energy X-Ray Imaging System: An Observer Preference Study in Healthy Volunteers.J Clin Med. 2025 Mar 19;14(6):2091. doi: 10.3390/jcm14062091. J Clin Med. 2025. PMID: 40142899 Free PMC article.
References
-
- Koh H.K., Geller A.C., VanderWeele T.J. Deaths from COVID-19. JAMA. 2021;325(2):133–134. - PubMed
-
- Shirani F., Shayganfar A., Hajiahmadi S. COVID-19 pneumonia: a pictorial review of CT findings and differential diagnosis. Egypt. J. Radiol. Nucl. Med. 2021;52(1):1–8.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

