Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients
- PMID: 36208629
- PMCID: PMC9510054
- DOI: 10.1016/j.xcrm.2022.100779
Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients
Abstract
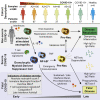
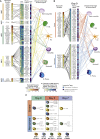
Mechanisms of neutrophil involvement in severe coronavirus disease 2019 (COVID-19) remain incompletely understood. Here, we collect longitudinal blood samples from 306 hospitalized COVID-19+ patients and 86 controls and perform bulk RNA sequencing of enriched neutrophils, plasma proteomics, and high-throughput antibody profiling to investigate relationships between neutrophil states and disease severity. We identify dynamic switches between six distinct neutrophil subtypes. At days 3 and 7 post-hospitalization, patients with severe disease display a granulocytic myeloid-derived suppressor cell-like gene expression signature, while patients with resolving disease show a neutrophil progenitor-like signature. Humoral responses are identified as potential drivers of neutrophil effector functions, with elevated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific immunoglobulin G1 (IgG1)-to-IgA1 ratios in plasma of severe patients who survived. In vitro experiments confirm that while patient-derived IgG antibodies induce phagocytosis in healthy donor neutrophils, IgA antibodies predominantly induce neutrophil cell death. Overall, our study demonstrates a dysregulated myelopoietic response in severe COVID-19 and a potential role for IgA-dominant responses contributing to mortality.
Keywords: COVID-19; G-MDSC; IgA; NETosis; SARS-CoV-2; degranulation; neutrophil; transcriptomics.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.S.-F. received funding from Bristol-Myers Squibb. G.A. is a founder of Seromyx Systems Inc. N.H. holds equity in Biontech and holds equity in and advises Danger Bio.
Figures






Update of
-
Longitudinal characterization of circulating neutrophils uncovers distinct phenotypes associated with disease severity in hospitalized COVID-19 patients.bioRxiv [Preprint]. 2021 Oct 5:2021.10.04.463121. doi: 10.1101/2021.10.04.463121. bioRxiv. 2021. Update in: Cell Rep Med. 2022 Oct 18;3(10):100779. doi: 10.1016/j.xcrm.2022.100779. PMID: 34642692 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

