Delivering genes with human immunodeficiency virus-derived vehicles: still state-of-the-art after 25 years
- PMID: 36209077
- PMCID: PMC9548131
- DOI: 10.1186/s12929-022-00865-4
Delivering genes with human immunodeficiency virus-derived vehicles: still state-of-the-art after 25 years
Abstract
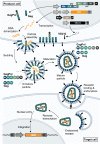
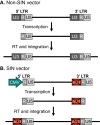
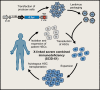
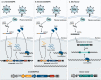
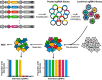
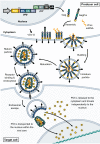
Viruses are naturally endowed with the capacity to transfer genetic material between cells. Following early skepticism, engineered viruses have been used to transfer genetic information into thousands of patients, and genetic therapies are currently attracting large investments. Despite challenges and severe adverse effects along the way, optimized technologies and improved manufacturing processes are driving gene therapy toward clinical translation. Fueled by the outbreak of AIDS in the 1980s and the accompanying focus on human immunodeficiency virus (HIV), lentiviral vectors derived from HIV have grown to become one of the most successful and widely used vector technologies. In 2022, this vector technology has been around for more than 25 years. Here, we celebrate the anniversary by portraying the vector system and its intriguing properties. We dive into the technology itself and recapitulate the use of lentiviral vectors for ex vivo gene transfer to hematopoietic stem cells and for production of CAR T-cells. Furthermore, we describe the adaptation of lentiviral vectors for in vivo gene delivery and cover the important contribution of lentiviral vectors to basic molecular research including their role as carriers of CRISPR genome editing technologies. Last, we dwell on the emerging capacity of lentiviral particles to package and transfer foreign proteins.
Keywords: CRISPR; Gene editing; Gene therapy; Genome engineering; HIV; IDLV; Integrase-defective lentiviral vectors; Lentiviral vectors; Lentivirus.
© 2022. The Author(s).
Conflict of interest statement
No competing financial interests exist.
Figures




References
-
- Centers for Disease C. Pneumocystis pneumonia--Los Angeles. MMWR Morb Mortal Wkly Rep. 1981;30(21):250–2. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

