Pivotal interplays between fecal metabolome and gut microbiome reveal functional signatures in cerebral ischemic stroke
- PMID: 36209079
- PMCID: PMC9548195
- DOI: 10.1186/s12967-022-03669-0
Pivotal interplays between fecal metabolome and gut microbiome reveal functional signatures in cerebral ischemic stroke
Abstract
Background: Integrative analysis approaches of metagenomics and metabolomics have been widely developed to understand the association between disease and the gut microbiome. However, the different profiling patterns of different metabolic samples in the association analysis make it a matter of concern which type of sample is the most closely associated with gut microbes and disease. To address this lack of knowledge, we investigated the association between the gut microbiome and metabolomic profiles of stool, urine, and plasma samples from ischemic stroke patients and healthy subjects.
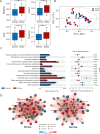
Methods: We performed metagenomic sequencing (feces) and untargeted metabolomics analysis (feces, plasma, and urine) from ischemic stroke patients and healthy volunteers. Differential analyses were conducted to find key differential microbiota and metabolites for ischemic stroke. Meanwhile, Spearman's rank correlation and linear regression analyses were used to study the association between microbiota and metabolites of different metabolic mixtures.
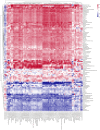
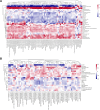
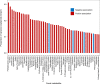
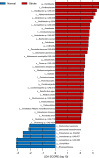
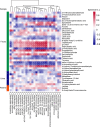
Results: Untargeted metabolomics analysis shows that feces had the most abundant features and identified metabolites, followed by urine and plasma. Feces had the highest number of differential metabolites between ischemic stroke patients and the healthy group. Based on the association analysis between metagenomics and metabolomics of fecal, urine, and plasma, fecal metabolome showed the strongest association with the gut microbiome. There are 1073, 191, and 81 statistically significant pairs (P < 0.05) in the correlation analysis for fecal, urine, and plasma metabolome. Fecal metabolites explained the variance of alpha-diversity of the gut microbiome up to 31.1%, while urine and plasma metabolites only explained the variance of alpha-diversity up to 13.5% and 10.6%. Meanwhile, there were more significant differential metabolites in feces than urine and plasma associated with the stroke marker bacteria.
Conclusions: The systematic association analysis between gut microbiome and metabolomics reveals that fecal metabolites show the strongest association with the gut microbiome, followed by urine and plasma. The findings would promote the association study between the gut microbiome and fecal metabolome to explore key factors that are associated with diseases. We also provide a user-friendly web server and a R package to facilitate researchers to conduct the association analysis of gut microbiome and metabolomics.
Keywords: Gut microbiota; Integrative analysis; Ischemic stroke; Metabolomics; Microbiome.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Wang H, Gou W, Su C, Du W, Zhang J, Miao Z, et al. Association of gut microbiota with glycaemic traits and incident type 2 diabetes, and modulation by habitual diet: a population-based longitudinal cohort study in Chinese adults. Diabetologia. 2022;65(9):1572. doi: 10.1007/s00125-022-05737-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

