Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management
- PMID: 36209487
- PMCID: PMC9759771
- DOI: 10.1002/cac2.12366
Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management
Abstract
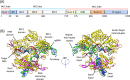
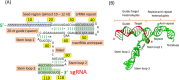
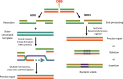
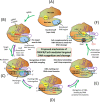
Clustered regularly interspaced short palindromic repeats-associated protein (CRISPR/Cas9), an adaptive microbial immune system, has been exploited as a robust, accurate, efficient and programmable method for genome targeting and editing. This innovative and revolutionary technique can play a significant role in animal modeling, in vivo genome therapy, engineered cell therapy, cancer diagnosis and treatment. The CRISPR/Cas9 endonuclease system targets a specific genomic locus by single guide RNA (sgRNA), forming a heteroduplex with target DNA. The Streptococcus pyogenes Cas9/sgRNA:DNA complex reveals a bilobed architecture with target recognition and nuclease lobes. CRISPR/Cas9 assembly can be hijacked, and its nanoformulation can be engineered as a delivery system for different clinical utilizations. However, the efficient and safe delivery of the CRISPR/Cas9 system to target tissues and cancer cells is very challenging, limiting its clinical utilization. Viral delivery strategies of this system may have many advantages, but disadvantages such as immune system stimulation, tumor promotion risk and small insertion size outweigh these advantages. Thus, there is a desperate need to develop an efficient non-viral physical delivery system based on simple nanoformulations. The delivery strategies of CRISPR/Cas9 by a nanoparticle-based system have shown tremendous potential, such as easy and large-scale production, combination therapy, large insertion size and efficient in vivo applications. This review aims to provide in-depth updates on Streptococcus pyogenic CRISPR/Cas9 structure and its mechanistic understanding. In addition, the advances in its nanoformulation-based delivery systems, including lipid-based, polymeric structures and rigid NPs coupled to special ligands such as aptamers, TAT peptides and cell-penetrating peptides, are discussed. Furthermore, the clinical applications in different cancers, clinical trials and future prospects of CRISPR/Cas9 delivery and genome targeting are also discussed.
Keywords: CRISPR/Cas9; cancer; clinical trials; genome editing; mechanism; nanoparticles; structure.
© 2022 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
The authors declare no conflict of interest
Figures

Similar articles
-
Innovative Strategies of Reprogramming Immune System Cells by Targeting CRISPR/Cas9-Based Genome-Editing Tools: A New Era of Cancer Management.Int J Nanomedicine. 2023 Sep 29;18:5531-5559. doi: 10.2147/IJN.S424872. eCollection 2023. Int J Nanomedicine. 2023. PMID: 37795042 Free PMC article. Review.
-
Recent Updates of the CRISPR/Cas9 Genome Editing System: Novel Approaches to Regulate Its Spatiotemporal Control by Genetic and Physicochemical Strategies.Int J Nanomedicine. 2024 Jun 6;19:5335-5363. doi: 10.2147/IJN.S455574. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38859956 Free PMC article. Review.
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: A Future Perspective of Cancer Management.Int J Mol Sci. 2023 Apr 11;24(8):7052. doi: 10.3390/ijms24087052. Int J Mol Sci. 2023. PMID: 37108214 Free PMC article. Review.
-
Recent Advances in Understanding the Molecular Mechanisms of Multidrug Resistance and Novel Approaches of CRISPR/Cas9-Based Genome-Editing to Combat This Health Emergency.Int J Nanomedicine. 2024 Feb 7;19:1125-1143. doi: 10.2147/IJN.S453566. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38344439 Free PMC article. Review.
Cited by
-
Innovative Strategies of Reprogramming Immune System Cells by Targeting CRISPR/Cas9-Based Genome-Editing Tools: A New Era of Cancer Management.Int J Nanomedicine. 2023 Sep 29;18:5531-5559. doi: 10.2147/IJN.S424872. eCollection 2023. Int J Nanomedicine. 2023. PMID: 37795042 Free PMC article. Review.
-
Leveraging protein language models for cross-variant CRISPR/Cas9 sgRNA activity prediction.Bioinformatics. 2025 Jul 1;41(7):btaf385. doi: 10.1093/bioinformatics/btaf385. Bioinformatics. 2025. PMID: 40600900 Free PMC article.
-
Tumor biomarkers for diagnosis, prognosis and targeted therapy.Signal Transduct Target Ther. 2024 May 20;9(1):132. doi: 10.1038/s41392-024-01823-2. Signal Transduct Target Ther. 2024. PMID: 38763973 Free PMC article. Review.
-
Nanotechnology in oncology: advances in biosynthesis, drug delivery, and theranostics.Discov Oncol. 2025 Jun 21;16(1):1172. doi: 10.1007/s12672-025-02664-3. Discov Oncol. 2025. PMID: 40542991 Free PMC article. Review.
-
Biomaterials-mediated CRISPR/Cas9 delivery: recent challenges and opportunities in gene therapy.Front Chem. 2023 Sep 28;11:1259435. doi: 10.3389/fchem.2023.1259435. eCollection 2023. Front Chem. 2023. PMID: 37841202 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

