SDCBP-AS1 destabilizes β-catenin by regulating ubiquitination and SUMOylation of hnRNP K to suppress gastric tumorigenicity and metastasis
- PMID: 36209503
- PMCID: PMC9648392
- DOI: 10.1002/cac2.12367
SDCBP-AS1 destabilizes β-catenin by regulating ubiquitination and SUMOylation of hnRNP K to suppress gastric tumorigenicity and metastasis
Abstract
Background: Gastric cancer (GC) is among the most malignant tumors, yet the pathogenesis is not fully understood, especially the lack of detailed information about the mechanisms underlying long non-coding RNA (lncRNA)-mediated post-translational modifications. Here, the molecular mechanisms and clinical significance of the novel lncRNA syndecan-binding protein 2-antisense RNA 1 (SDCBP2-AS1) in the tumorigenesis and progression of GC were investigated.
Methods: The expression levels of SDCBP2-AS1 in 132 pairs of GC and adjacent normal tissues were compared, and the biological functions were assessed in vitro and in vivo. RNA pull-down and immunoprecipitation assays were conducted to clarify the interactions of SDCBP2-AS1 and heterogeneous nuclear ribonucleoprotein (hnRNP) K. RNA-sequencing, immunoprecipitation, immunofluorescence, and luciferase analyses were performed to investigate the functions of SDCBP2-AS1.
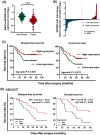
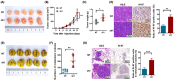
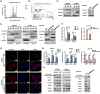
Results: SDCBP2-AS1 was significantly downregulated in GC tissues and predictive of poor patient prognosis. Silencing of SDCBP2-AS1 promoted the proliferation and migration of GC cells both in vitro and in vivo. Mechanically, SDCBP2-AS1 physically bound to hnRNP K to repress SUMOylation of hnRNP K and facilitated ubiquitination of hnRNP K and β-catenin, thereby promoting the degradation of β-catenin in the cytoplasm. Silencing of SDCBP2-AS1 caused SUMOylation of hnRNP K and stabilized β-catenin activity, which altered transcription of downstream genes, resulting in tumorigenesis and metastasis of GC. Moreover, the knockdown of hnRNP K partially abrogated the effects of SDCBP2-AS1.
Conclusions: SDCBP2-AS1 interacts with hnRNP K to suppress tumorigenesis and metastasis of GC and regulates post-transcriptional modifications of hnRNP K to destabilize β-catenin. These findings suggest SDCBP2-AS1 as a potential target for the treatment of GC.
Keywords: SDCBP2-AS1; gastric cancer; hnRNP K; post-transcriptional modifications; tumorigenesis; β-catenin.
© 2022 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





Similar articles
-
Highly expressed B3GALT5-AS1 contributes to gastric cancer progression by recruiting WDR5 to mediate B3GALT5 and regulating β-catenin/ZEB1 axis.J Cell Mol Med. 2024 Sep;28(17):e70061. doi: 10.1111/jcmm.70061. J Cell Mol Med. 2024. PMID: 39224045 Free PMC article.
-
Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA.Mol Cancer. 2022 Aug 19;21(1):168. doi: 10.1186/s12943-022-01638-1. Mol Cancer. 2022. PMID: 35986274 Free PMC article.
-
Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer.J Cell Mol Med. 2018 Oct;22(10):4751-4759. doi: 10.1111/jcmm.13726. Epub 2018 Jul 14. J Cell Mol Med. 2018. PMID: 30006956 Free PMC article.
-
Silencing of long non-coding RNA SDCBP2-AS1/microRNA-656-3p/CRIM1 axis promotes ferroptosis of lung cancer cells.Cell Mol Biol (Noisy-le-grand). 2023 Sep 30;69(9):189-194. doi: 10.14715/cmb/2023.69.9.29. Cell Mol Biol (Noisy-le-grand). 2023. PMID: 37807311
-
Implication of protein post translational modifications in gastric cancer.Front Cell Dev Biol. 2025 Feb 4;13:1523958. doi: 10.3389/fcell.2025.1523958. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39968176 Free PMC article. Review.
Cited by
-
LINC00629, a HOXB4-downregulated long noncoding RNA, inhibits glycolysis and ovarian cancer progression by destabilizing c-Myc.Cancer Sci. 2024 Mar;115(3):804-819. doi: 10.1111/cas.16049. Epub 2024 Jan 5. Cancer Sci. 2024. PMID: 38182548 Free PMC article.
-
The long noncoding RNA ELFN1-AS1 promotes gastric cancer growth and metastasis by interacting with TAOK1 to inhibit the Hippo signaling pathway.Cell Death Discov. 2024 Nov 11;10(1):465. doi: 10.1038/s41420-024-02235-5. Cell Death Discov. 2024. PMID: 39528458 Free PMC article.
-
CircERC1 facilitates chemoresistance through inhibiting pyroptosis and remodeling extracellular matrix in pancreatic cancer.Mol Cancer. 2025 Jul 2;24(1):185. doi: 10.1186/s12943-025-02385-9. Mol Cancer. 2025. PMID: 40605033 Free PMC article.
-
Distinctive tumorigenic significance and innovative oncology targets of SUMOylation.Theranostics. 2024 May 19;14(8):3127-3149. doi: 10.7150/thno.97162. eCollection 2024. Theranostics. 2024. PMID: 38855173 Free PMC article. Review.
-
Regulation of SUMOylation on RNA metabolism in cancers.Front Mol Biosci. 2023 Feb 24;10:1137215. doi: 10.3389/fmolb.2023.1137215. eCollection 2023. Front Mol Biosci. 2023. PMID: 36911524 Free PMC article. Review.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209‐49. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

