Structural modeling of protein ensembles between E3 RING ligases and SARS-CoV-2: The role of zinc binding domains
- PMID: 36209710
- PMCID: PMC9531365
- DOI: 10.1016/j.jtemb.2022.127089
Structural modeling of protein ensembles between E3 RING ligases and SARS-CoV-2: The role of zinc binding domains
Abstract
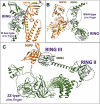
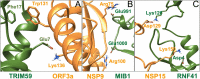
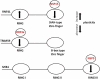
Background: The ubiquitin system is a modification process with many different cellular functions including immune signaling and antiviral functions. E3 ubiquitin ligases are enzymes that recruit an E2 ubiquitin-conjugating enzyme bound to ubiquitin in order to catalyze the transfer of ubiquitin from the E2 to a protein substrate. The RING E3s, the most abundant type of ubiquitin ligases, are characterized by a zinc (II)-binding domain called RING (Really Interesting New Gene). Viral replication requires modifying and hijacking key cellular pathways within host cells such as cellular ubiquitination. There are well-established examples where a viral proteins bind to RING E3s, redirecting them to degrade otherwise long-lived host proteins or inhibiting E3's ubiquitination activity. Recently, three binary interactions between SARS-CoV-2 proteins and innate human immune signaling Ε3 RING ligases: NSP15-RNF41, ORF3a-TRIM59 and NSP9-MIB1 have been experimentally established.
Methods: In this work, we have investigated the mode of the previous experimentally supported NSP15-RNF41, ORF3a,-TRIM59 and NSP9-MIB1 binary interactions by in silico methodologies intending to provide structural insights of E3-virus interplay that can help identify potential inhibitors that could block SARS-CoV-2 infection of immune cells.
Conclusion: In silico methodologies have shown that the above human E3 ligases interact with viral partners through their Zn(II) binding domains. This RING mediated formation of stable SARS-CoV-2-E3 complexes indicates a critical structural role of RING domains in immune system disruption by SARS-CoV-2-infection.
Data availability: The data used to support the findings of this research are included within the article and are labeled with references.
Keywords: E3 RING ligases; Haddock; SARS-CoV-2; Zinc binding RING domains.
Copyright © 2022 Elsevier GmbH. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no potential conflicts of interest.
Figures

References
-
- Shemesh M., Aktepe T.E., Deerain J.M., McAuley J.L., Audsley M.D., David C.T., Purcell D.F.J., Urin V., Hartmann R., Moseley G.W., Mackenzie J.M., Schreiber G., Harari D. SARS-CoV-2 suppresses IFNbeta production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLOS Pathog. 2021;17(8) doi: 10.1371/journal.ppat.1009800. - DOI - PMC - PubMed
-
- Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., Ran X., Damani-Yokota P., Tang H., Karakousi T., Son J., Kaczmarek M.E., Zhang Z., Yeung S.T., McCune B.T., Chen R.E., Tang F., Ren X., Chen X., Hsu J.C.C., Teplova M., Huang B., Deng H., Long Z., Mudianto T., Jin S., Lin P., Du J., Zang R., Su T.T., Herrera A., Zhou M., Yan R., Cui J., Zhu J., Zhou Q., Wang T., Ma J., Koralov S.B., Aifantis I., Segal L.N., Diamond M.S., Khanna K.M., Stapleford K.A., Cresswell P., Liu Y., Ding S., Xie Q., Wang J. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54(6):1304–1319. doi: 10.1016/j.immuni.2021.05.006. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

