Copy Number Variants Are Ovarian Cancer Risk Alleles at Known and Novel Risk Loci
- PMID: 36210504
- PMCID: PMC9949586
- DOI: 10.1093/jnci/djac160
Copy Number Variants Are Ovarian Cancer Risk Alleles at Known and Novel Risk Loci
Abstract
Background: Known risk alleles for epithelial ovarian cancer (EOC) account for approximately 40% of the heritability for EOC. Copy number variants (CNVs) have not been investigated as EOC risk alleles in a large population cohort.
Methods: Single nucleotide polymorphism array data from 13 071 EOC cases and 17 306 controls of White European ancestry were used to identify CNVs associated with EOC risk using a rare admixture maximum likelihood test for gene burden and a by-probe ratio test. We performed enrichment analysis of CNVs at known EOC risk loci and functional biofeatures in ovarian cancer-related cell types.
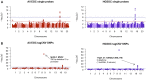
Results: We identified statistically significant risk associations with CNVs at known EOC risk genes; BRCA1 (PEOC = 1.60E-21; OREOC = 8.24), RAD51C (Phigh-grade serous ovarian cancer [HGSOC] = 5.5E-4; odds ratio [OR]HGSOC = 5.74 del), and BRCA2 (PHGSOC = 7.0E-4; ORHGSOC = 3.31 deletion). Four suggestive associations (P < .001) were identified for rare CNVs. Risk-associated CNVs were enriched (P < .05) at known EOC risk loci identified by genome-wide association study. Noncoding CNVs were enriched in active promoters and insulators in EOC-related cell types.
Conclusions: CNVs in BRCA1 have been previously reported in smaller studies, but their observed frequency in this large population-based cohort, along with the CNVs observed at BRCA2 and RAD51C gene loci in EOC cases, suggests that these CNVs are potentially pathogenic and may contribute to the spectrum of disease-causing mutations in these genes. CNVs are likely to occur in a wider set of susceptibility regions, with potential implications for clinical genetic testing and disease prevention.
© The Author(s) 2022. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
Grants and funding
- P50 CA159981/CA/NCI NIH HHS/United States
- UM1 CA176726/CA/NCI NIH HHS/United States
- R01 CA058860/CA/NCI NIH HHS/United States
- P50 CA105009/CA/NCI NIH HHS/United States
- R01-CA122443/NH/NIH HHS/United States
- 076113/WT_/Wellcome Trust/United Kingdom
- MC_UU_00004/01/MRC_/Medical Research Council/United Kingdom
- P30 CA015083/CA/NCI NIH HHS/United States
- N01 CN025403/CA/NCI NIH HHS/United States
- P01 CA017054/CA/NCI NIH HHS/United States
- MR/M012190/1/MRC_/Medical Research Council/United Kingdom
- C490/A10119 C490/A10124/CRUK_/Cancer Research UK/United Kingdom
- 1000143/MRC_/Medical Research Council/United Kingdom
- R01-CA54419/NH/NIH HHS/United States
- C8221/A19170/CRUK_/Cancer Research UK/United Kingdom
- R01 CA049449/CA/NCI NIH HHS/United States
- T32 GM118288/GM/NIGMS NIH HHS/United States
- CA1X01HG007491-01/NH/NIH HHS/United States
- Z01-ES044005/ES/NIEHS NIH HHS/United States
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- R01 CA106414/CA/NCI NIH HHS/United States
- R01 CA095023/CA/NCI NIH HHS/United States
- N01 PC067010/PC/NCI NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- R01 CA058598/CA/NCI NIH HHS/United States
- U01 CA176726/CA/NCI NIH HHS/United States
- Z01 ES049033/ImNIH/Intramural NIH HHS/United States
- S10 RR025141/RR/NCRR NIH HHS/United States
- M01 RR000056/RR/NCRR NIH HHS/United States
- DH_/Department of Health/United Kingdom
- G1000143/MRC_/Medical Research Council/United Kingdom
- 5T32GM118288-03/NH/NIH HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- K07-CA080668/BC/NCI NIH HHS/United States
- 14136 /CRUK_/Cancer Research UK/United Kingdom
- AICR_/Worldwide Cancer Research/United Kingdom
- U19-CA148112/CA/NCI NIH HHS/United States
- G0401527/MRC_/Medical Research Council/United Kingdom
- MR_UU_12023/MRC_/Medical Research Council/United Kingdom
- R01 CA067262/CA/NCI NIH HHS/United States
- UM1 CA186107/CA/NCI NIH HHS/United States
- R01 CA076016/CA/NCI NIH HHS/United States
- HG/NHGRI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- R01- CA61107/CA/NCI NIH HHS/United States
- R01-CA58598/NH/NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- ULTR000445/TR/NCATS NIH HHS/United States
- R03 CA115195/CA/NCI NIH HHS/United States
- BBC_/Breast Cancer Now/United Kingdom
- R01 CA160669/CA/NCI NIH HHS/United States
- R01-CA058860/NH/NIH HHS/United States
- C570/A16491/CRUK_/Cancer Research UK/United Kingdom
- R01-CA76016/NH/NIH HHS/United States
- R01-CA106414-A2/NH/NIH HHS/United States
- 001/WHO_/World Health Organization/International
- P50 CA136393/CA/NCI NIH HHS/United States
- R01 CA126841/CA/NCI NIH HHS/United States
- 209057/WT_/Wellcome Trust/United Kingdom
- R03 CA113148/CA/NCI NIH HHS/United States
- R01 CA149429/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

