Click chemistry functionalization of self-assembling peptide hydrogels
- PMID: 36210776
- PMCID: PMC10092743
- DOI: 10.1002/jbm.a.37460
Click chemistry functionalization of self-assembling peptide hydrogels
Abstract
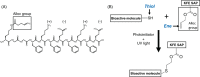
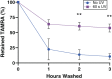
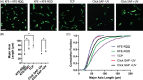
Self-assembling peptide (SAP) hydrogels provide a fibrous microenvironment to cells while also giving users control of biochemical and mechanical cues. Previously, biochemical cues were introduced by physically mixing them with SAPs prior to hydrogel assembly, or by incorporating them into the SAP sequence during peptide synthesis, which limited flexibility and increased costs. To circumvent these limitations, we developed "Click SAPs," a novel formulation that can be easily functionalized via click chemistry thiol-ene reaction. Due to its high cytocompatibility, the thiol-ene click reaction is currently used to crosslink and functionalize other types of polymeric hydrogels. In this study, we developed a click chemistry compatible SAP platform by addition of a modified lysine (lysine-alloc) to the SAP sequence, enabling effective coupling of thiol-containing molecules to the SAP hydrogel network. We demonstrate the flexibility of this approach by incorporating a fluorescent dye, a cellular adhesion peptide, and a matrix metalloproteinase-sensitive biosensor using the thiol-ene reaction in 3D Click SAPs. Using atomic force microscopy, we demonstrate that Click SAPs retain the ability to self-assemble into fibers, similar to previous systems. Additionally, a range of physiologically relevant stiffnesses can be achieved by adjusting SAP concentration. Encapsulated cells maintain high viability in Click SAPs and can interact with adhesion peptides and a matrix metalloproteinase biosensor, demonstrating that incorporated molecules retain their biological activity. The Click SAP platform supports easier functionalization with a wider array of bioactive molecules and enables new investigations with temporal and spatial control of the cellular microenvironment.
Keywords: click chemistry; extracellular matrix; hydrogel; matrix metalloproteinase; self-assembling peptide.
© 2022 The Authors. Journal of Biomedical Materials Research Part A published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Pérez CMR, Rank LA, Chmielewski J. Tuning the thermosensitive properties of hybrid collagen peptide–polymer hydrogels. Chem Commun. 2014;50(60):8174‐8176. - PubMed
-
- Hogrebe NJ, Reinhardt JW, Gooch KJ. Biomaterial microarchitecture: a potent regulator of individual cell behavior and multicellular organization. J Biomed Mater Res A. 2017;105(2):640‐661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

