A genome-wide association study of plasma concentrations of warfarin enantiomers and metabolites in sub-Saharan black-African patients
- PMID: 36210801
- PMCID: PMC9537548
- DOI: 10.3389/fphar.2022.967082
A genome-wide association study of plasma concentrations of warfarin enantiomers and metabolites in sub-Saharan black-African patients
Abstract
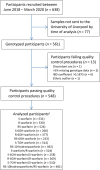
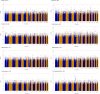
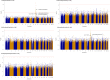
Diversity in pharmacogenomic studies is poor, especially in relation to the inclusion of black African patients. Lack of funding and difficulties in recruitment, together with the requirement for large sample sizes because of the extensive genetic diversity in Africa, are amongst the factors which have hampered pharmacogenomic studies in Africa. Warfarin is widely used in sub-Saharan Africa, but as in other populations, dosing is highly variable due to genetic and non-genetic factors. In order to identify genetic factors determining warfarin response variability, we have conducted a genome-wide association study (GWAS) of plasma concentrations of warfarin enantiomers/metabolites in sub-Saharan black-Africans. This overcomes the issue of non-adherence and may have greater sensitivity at genome-wide level, to identify pharmacokinetic gene variants than focusing on mean weekly dose, the usual end-point used in previous studies. Participants recruited at 12 outpatient sites in Uganda and South Africa on stable warfarin dose were genotyped using the Illumina Infinium H3Africa Consortium Array v2. Imputation was conducted using the 1,000 Genomes Project phase III reference panel. Warfarin/metabolite plasma concentrations were determined by high-performance liquid chromatography with tandem mass spectrometry. Multivariable linear regression was undertaken, with adjustment made for five non-genetic covariates and ten principal components of genetic ancestry. After quality control procedures, 548 participants and 17,268,054 SNPs were retained. CYP2C9*8, CYP2C9*9, CYP2C9*11, and the CYP2C cluster SNP rs12777823 passed the Bonferroni-adjusted replication significance threshold (p < 3.21E-04) for warfarin/metabolite ratios. In an exploratory GWAS analysis, 373 unique SNPs in 13 genes, including CYP2C9*8, passed the Bonferroni-adjusted genome-wide significance threshold (p < 3.846E-9), with 325 (87%, all located on chromosome 10) SNPs being associated with the S-warfarin/R-warfarin outcome (top SNP rs11188082, CYP2C19 intron variant, p = 1.55E-17). Approximately 69% of these SNPs were in linkage disequilibrium (r 2 > 0.8) with CYP2C9*8 (n = 216) and rs12777823 (n = 8). Using a pharmacokinetic approach, we have shown that variants other than CYP2C9*2 and CYP2C9*3 are more important in sub-Saharan black-Africans, mainly due to the allele frequencies. In exploratory work, we conducted the first warfarin pharmacokinetics-related GWAS in sub-Saharan Africans and identified novel SNPs that will require external replication and functional characterization before they can be considered for inclusion in warfarin dosing algorithms.
Keywords: black-African; genome-wide association study; personalized medicine; pharmacokinetics; warfarin.
Copyright © 2022 Asiimwe, Blockman, Cohen, Cupido, Hutchinson, Jacobson, Lamorde, Morgan, Mouton, Nakagaayi, Okello, Schapkaitz, Sekaggya-Wiltshire, Semakula, Waitt, Zhang, Jorgensen and Pirmohamed.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Local ancestry informed GWAS of warfarin dose requirement in African Americans identifies a novel CYP2C19 splice QTL.medRxiv [Preprint]. 2025 Mar 5:2025.03.03.25323247. doi: 10.1101/2025.03.03.25323247. medRxiv. 2025. PMID: 40093246 Free PMC article. Preprint.
-
Developing and Validating a Clinical Warfarin Dose-Initiation Model for Black-African Patients in South Africa and Uganda.Clin Pharmacol Ther. 2021 Jun;109(6):1564-1574. doi: 10.1002/cpt.2128. Epub 2020 Dec 28. Clin Pharmacol Ther. 2021. PMID: 33280090
-
Evaluation of Genome Wide Association Study Associated Type 2 Diabetes Susceptibility Loci in Sub Saharan Africans.Front Genet. 2015 Nov 24;6:335. doi: 10.3389/fgene.2015.00335. eCollection 2015. Front Genet. 2015. PMID: 26635871 Free PMC article.
-
Pharmacogenetics of warfarin elimination and its clinical implications.Clin Pharmacokinet. 2001;40(8):587-603. doi: 10.2165/00003088-200140080-00003. Clin Pharmacokinet. 2001. PMID: 11523725 Review.
-
Pharmacogenomics of CYP2C9: Functional and Clinical Considerations.J Pers Med. 2017 Dec 28;8(1):1. doi: 10.3390/jpm8010001. J Pers Med. 2017. PMID: 29283396 Free PMC article. Review.
Cited by
-
A meta-analysis and polygenic score study identifies novel genetic markers for waist-hip ratio in African populations.Obesity (Silver Spring). 2024 Nov;32(11):2175-2185. doi: 10.1002/oby.24123. Epub 2024 Oct 1. Obesity (Silver Spring). 2024. PMID: 39351966
-
LA-GEM: imputation of gene expression with incorporation of Local Ancestry.Pac Symp Biocomput. 2024;29:341-358. Pac Symp Biocomput. 2024. PMID: 38160291 Free PMC article.
-
Novel Gene Polymorphisms for Stable Warfarin Dose in a Korean Population: Genome-Wide Association Study.Biomedicines. 2023 Aug 19;11(8):2308. doi: 10.3390/biomedicines11082308. Biomedicines. 2023. PMID: 37626805 Free PMC article.
-
Meta-analysis of genome-wide association studies of stable warfarin dose in patients of African ancestry.Blood Adv. 2024 Oct 22;8(20):5248-5261. doi: 10.1182/bloodadvances.2024014227. Blood Adv. 2024. PMID: 39163621 Free PMC article.
-
Local ancestry informed GWAS of warfarin dose requirement in African Americans identifies a novel CYP2C19 splice QTL.medRxiv [Preprint]. 2025 Mar 5:2025.03.03.25323247. doi: 10.1101/2025.03.03.25323247. medRxiv. 2025. PMID: 40093246 Free PMC article. Preprint.
References
-
- Asiimwe I. G., Blockman M., Cohen K., Cupido C., Hutchinson C., Jacobson B., et al. (2021). Stable warfarin dose prediction in sub-saharan african patients: A machine-learning approach and external validation of a clinical dose-initiation algorithm. CPT. Pharmacometrics Syst. Pharmacol. 11, 20–29. 10.1002/psp4.12740 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

