Equilibrium-based COVID-19 diagnosis from routine blood tests: A sparse deep convolutional model
- PMID: 36210961
- PMCID: PMC9527205
- DOI: 10.1016/j.eswa.2022.118935
Equilibrium-based COVID-19 diagnosis from routine blood tests: A sparse deep convolutional model
Abstract
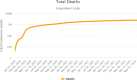
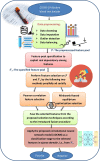
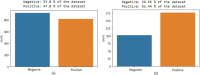
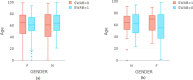
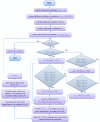
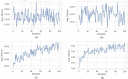
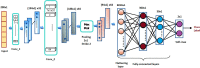
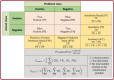
SARS-CoV2 (COVID-19) is the virus that causes the pandemic that has severely impacted human society with a massive death toll worldwide. Hence, there is a persistent need for fast and reliable automatic tools to help health teams in making clinical decisions. Predictive models could potentially ease the strain on healthcare systems by early and reliable screening of COVID-19 patients which helps to combat the spread of the disease. Recent studies have reported some key advantages of employing routine blood tests for initial screening of COVID-19 patients. Thus, in this paper, we propose a novel COVID-19 prediction model based on routine blood tests. In this model, we depend on exploiting the real dependency among the employed feature pool by a sparsification procedure. In this sparse domain, a hybrid feature selection mechanism is proposed. This mechanism fuses the selected features from two perspectives, the first is Pearson correlation and the second is a new Minkowski-based equilibrium optimizer (MEO). Then, the selected features are fed into a new 1D Convolutional Neural Network (1DCNN) for a final diagnosis decision. The proposed prediction model is tested with a new public dataset from San Raphael Hospital, Milan, Italy, i.e., OSR dataset which has two sub-datasets. According to the experimental results, the proposed model outperforms the state-of-the-art techniques with an average testing accuracy of 98.5% while we employ only less than half the size of the feature pool, i.e., we need only less than half the given blood tests in the employed dataset to get a final diagnosis decision.
Keywords: 1DCNN; Blood tests; COVID-19; Equilibrium optimization; Feature pool sparsification; Feature selection; Pearson correlation.
© 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

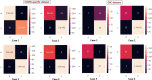
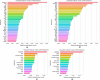
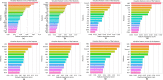

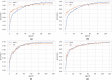

Similar articles
-
Artificial intelligence in clinical care amidst COVID-19 pandemic: A systematic review.Comput Struct Biotechnol J. 2021;19:2833-2850. doi: 10.1016/j.csbj.2021.05.010. Epub 2021 May 7. Comput Struct Biotechnol J. 2021. PMID: 34025952 Free PMC article. Review.
-
Hybrid optimal feature selection-based iterative deep convolution learning for COVID-19 classification system.Comput Biol Med. 2024 Oct;181:109031. doi: 10.1016/j.compbiomed.2024.109031. Epub 2024 Aug 21. Comput Biol Med. 2024. PMID: 39173484
-
OptCoNet: an optimized convolutional neural network for an automatic diagnosis of COVID-19.Appl Intell (Dordr). 2021;51(3):1351-1366. doi: 10.1007/s10489-020-01904-z. Epub 2020 Sep 21. Appl Intell (Dordr). 2021. PMID: 34764551 Free PMC article.
-
Human monkeypox disease prediction using novel modified restricted Boltzmann machine-based equilibrium optimizer.Sci Rep. 2024 Jul 30;14(1):17612. doi: 10.1038/s41598-024-68836-3. Sci Rep. 2024. PMID: 39080387 Free PMC article.
-
Universal screening for SARS-CoV-2 infection: a rapid review.Cochrane Database Syst Rev. 2020 Sep 15;9(9):CD013718. doi: 10.1002/14651858.CD013718. Cochrane Database Syst Rev. 2020. PMID: 33502003 Free PMC article.
Cited by
-
Recent Developments in Equilibrium Optimizer Algorithm: Its Variants and Applications.Arch Comput Methods Eng. 2023 Apr 12:1-54. doi: 10.1007/s11831-023-09923-y. Online ahead of print. Arch Comput Methods Eng. 2023. PMID: 37359743 Free PMC article.
-
SARS-CoV-2 Prediction Strategy Based on Classification Algorithms from a Full Blood Examination.ScientificWorldJournal. 2023 Aug 22;2023:3248192. doi: 10.1155/2023/3248192. eCollection 2023. ScientificWorldJournal. 2023. PMID: 37649715 Free PMC article.
References
-
- Alafif T., Tehame A.M., Bajaba S., Barnawi A., Zia S. Machine and deep learning towards COVID-19 diagnosis and treatment: Survey, challenges, and future directions. International Journal of Environmental Research and Public Health. 2021;18(3):1117. doi: 10.31224/osf.io/w3zxy. - DOI - PMC - PubMed
-
- Albahri, O. S., Zaidan, A. A., Albahri, A. S., Zaidan, B. B., Abdulkareem, K. H., Al-Qaysi, Z. T., ... & Rashid, N. A. (2020). Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: Taxonomy analysis, challenges, future solutions and methodological aspects. Journal of Infection and Public Health, 13(10), 1381-1396.https://doi.org/10.1016/j.jiph.2020.06.028. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
