Maternal gut microbiota during pregnancy and the composition of immune cells in infancy
- PMID: 36211431
- PMCID: PMC9535361
- DOI: 10.3389/fimmu.2022.986340
Maternal gut microbiota during pregnancy and the composition of immune cells in infancy
Abstract
Background: Preclinical studies have shown that maternal gut microbiota during pregnancy play a key role in prenatal immune development but the relevance of these findings to humans is unknown. The aim of this prebirth cohort study was to investigate the association between the maternal gut microbiota in pregnancy and the composition of the infant's cord and peripheral blood immune cells over the first year of life.
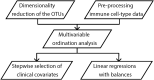
Methods: The Barwon Infant Study cohort (n=1074 infants) was recruited using an unselected sampling frame. Maternal fecal samples were collected at 36 weeks of pregnancy and flow cytometry was conducted on cord/peripheral blood collected at birth, 6 and 12 months of age. Among a randomly selected sub-cohort with available samples (n=293), maternal gut microbiota was characterized by sequencing the 16S rRNA V4 region. Operational taxonomic units (OTUs) were clustered based on their abundance. Associations between maternal fecal microbiota clusters and infant granulocyte, monocyte and lymphocyte subsets were explored using compositional data analysis. Partial least squares (PLS) and regression models were used to investigate the relationships/associations between environmental, maternal and infant factors, and OTU clusters.
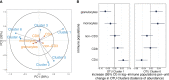
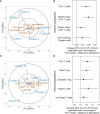
Results: We identified six clusters of co-occurring OTUs. The first two components in the PLS regression explained 39% and 33% of the covariance between the maternal prenatal OTU clusters and immune cell populations in offspring at birth. A cluster in which Dialister, Escherichia, and Ruminococcus were predominant was associated with a lower proportion of granulocytes (p=0.002), and higher proportions of both central naïve CD4+ T cells (CD4+/CD45RA+/CD31-) (p<0.001) and naïve regulatory T cells (Treg) (CD4+/CD45RA+/FoxP3low) (p=0.02) in cord blood. The association with central naïve CD4+ T cells persisted to 12 months of age.
Conclusion: This birth cohort study provides evidence consistent with past preclinical models that the maternal gut microbiota during pregnancy plays a role in shaping the composition of innate and adaptive elements of the infant's immune system following birth.
Keywords: birth cohort; fetal immunity; gut microbiota; maternal microbiota; neonatal T cells.
Copyright © 2022 Gao, O’Hely, Quinn, Ponsonby, Harrison, Frøkiær, Tang, Brix, Kristiansen, Burgner, Saffery, Ranganathan, Collier and Vuillermin.
Conflict of interest statement
The findings described in this paper are the subject of a provisional patent, licensed to Prevatex Pty Ltd, in which the following authors have a financial interest: PV, MOH, SR, ALP. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner.Microbiome. 2018 Jul 5;6(1):109. doi: 10.1186/s40168-018-0490-8. Microbiome. 2018. PMID: 29973274 Free PMC article.
-
Does birth mode modify associations of maternal pre-pregnancy BMI and gestational weight gain with the infant gut microbiome?Int J Obes (Lond). 2020 Jan;44(1):23-32. doi: 10.1038/s41366-018-0273-0. Epub 2019 Feb 14. Int J Obes (Lond). 2020. PMID: 30765892 Free PMC article.
-
Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants.Microbiome. 2017 Sep 4;5(1):113. doi: 10.1186/s40168-017-0332-0. Microbiome. 2017. PMID: 28870230 Free PMC article.
-
Fetal programming of overweight through the microbiome: boys are disproportionately affected.J Dev Orig Health Dis. 2016 Feb;7(1):25-34. doi: 10.1017/S2040174415001269. Epub 2015 Jun 29. J Dev Orig Health Dis. 2016. PMID: 26118444 Review.
-
Maternal vitamin D in pregnancy and infant's gut microbiota: a systematic review.Front Pediatr. 2023 Oct 16;11:1248517. doi: 10.3389/fped.2023.1248517. eCollection 2023. Front Pediatr. 2023. PMID: 37915988 Free PMC article. Review.
Cited by
-
Higher gut Bacteroidetes and Actinobacteria population in early pregnancy is associated with lower risk of gestational diabetes in the second trimester.BMC Pregnancy Childbirth. 2025 Feb 3;25(1):106. doi: 10.1186/s12884-025-07192-0. BMC Pregnancy Childbirth. 2025. PMID: 39901086 Free PMC article.
-
Influenza A virus infection during pregnancy causes immunological changes in gut-associated lymphoid tissues of offspring mice.Am J Physiol Gastrointest Liver Physiol. 2023 Sep 1;325(3):G230-G238. doi: 10.1152/ajpgi.00062.2023. Epub 2023 Jul 11. Am J Physiol Gastrointest Liver Physiol. 2023. PMID: 37431584 Free PMC article.
-
A maternal sweet diet is associated with the gut dysbiosis in the first trimester of pregnancy.BMC Nutr. 2024 Dec 18;10(1):162. doi: 10.1186/s40795-024-00972-5. BMC Nutr. 2024. PMID: 39695908 Free PMC article.
-
The Effects of Fecal Microbial Transplantation on the Symptoms in Autism Spectrum Disorder, Gut Microbiota and Metabolites: A Scoping Review.Microorganisms. 2025 May 31;13(6):1290. doi: 10.3390/microorganisms13061290. Microorganisms. 2025. PMID: 40572178 Free PMC article. Review.
-
Gut microbiota influences the efficiency of immune checkpoint inhibitors by modulating the immune system (Review).Oncol Lett. 2024 Jan 5;27(2):87. doi: 10.3892/ol.2024.14221. eCollection 2024 Feb. Oncol Lett. 2024. PMID: 38249807 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

