ZAR1: Guardian of plant kinases
- PMID: 36212348
- PMCID: PMC9539561
- DOI: 10.3389/fpls.2022.981684
ZAR1: Guardian of plant kinases
Abstract
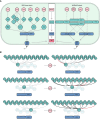
A key facet of innate immunity in plants entails the recognition of pathogen "effector" virulence proteins by host Nucleotide-Binding Leucine-Rich Repeat Receptors (NLRs). Among characterized NLRs, the broadly conserved ZAR1 NLR is particularly remarkable due to its capacity to recognize at least six distinct families of effectors from at least two bacterial genera. This expanded recognition spectrum is conferred through interactions between ZAR1 and a dynamic network of two families of Receptor-Like Cytoplasmic Kinases (RLCKs): ZED1-Related Kinases (ZRKs) and PBS1-Like Kinases (PBLs). In this review, we survey the history of functional studies on ZAR1, with an emphasis on how the ZAR1-RLCK network functions to trap diverse effectors. We discuss 1) the dynamics of the ZAR1-associated RLCK network; 2) the specificity between ZRKs and PBLs; and 3) the specificity between effectors and the RLCK network. We posit that the shared protein fold of kinases and the switch-like properties of their interactions make them ideal effector sensors, enabling ZAR1 to act as a broad spectrum guardian of host kinases.
Keywords: PBL; ZAR1; ZRK; arabidopsis; effector-triggered immunity; kinases; network; pseudokinases.
Copyright © 2022 Breit-McNally, Laflamme, Singh, Desveaux and Guttman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

