Longitudinal analysis of exposure to a low concentration of oxytetracycline on the zebrafish gut microbiome
- PMID: 36212820
- PMCID: PMC9536460
- DOI: 10.3389/fmicb.2022.985065
Longitudinal analysis of exposure to a low concentration of oxytetracycline on the zebrafish gut microbiome
Abstract
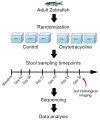
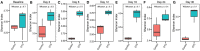
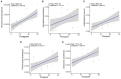
Oxytetracycline, a widely produced and administered antibiotic, is uncontrollably released in low concentrations in various types of environments. However, the impact of exposure to such low concentrations of antibiotics on the host remains poorly understood. In this study, we exposed zebrafish to a low concentration (5,000 ng/L) of oxytetracycline for 1 month, collected samples longitudinally (Baseline, and Days 3, 6, 9, 12, 24, and 30), and elucidated the impact of exposure on microbial composition, antibiotic resistance genes, mobile genetic elements, and phospholipid metabolism pathway through comparison of the sequenced data with respective sequence databases. We identified Pseudomonas aeruginosa, a well-known pathogen, to be significantly positively associated with the duration of oxytetracycline exposure (Adjusted P = 5.829e-03). Several tetracycline resistance genes (e.g., tetE) not only showed significantly higher abundance in the exposed samples but were also positively associated with the duration of exposure (Adjusted P = 1.114e-02). Furthermore, in the exposed group, the relative abundance of genes involved in phospholipid metabolism had also decreased. Lastly, we characterized the impact of exposure on zebrafish intestinal structure and found that the goblet cell counts were decreased (~82%) after exposure. Overall, our results show that a low concentration of oxytetracycline can increase the abundance of pathogenic bacteria and lower the abundance of key metabolic pathways in the zebrafish gut microbiome that can render them prone to bacterial infections and health-associated complications.
Keywords: antibiotic exposure; environmental pollution; goblet cells; gut microbiome; longitudinal analysis; low concentration; oxytetracycline.
Copyright © 2022 Kayani, Yu, Qiu, Yu, Chen and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Environmental concentrations of antibiotics alter the zebrafish gut microbiome structure and potential functions.Environ Pollut. 2021 Jun 1;278:116760. doi: 10.1016/j.envpol.2021.116760. Epub 2021 Feb 22. Environ Pollut. 2021. PMID: 33725532
-
Mite gut microbiome and resistome exhibited species-specific and dose-dependent effect in response to oxytetracycline exposure.Sci Total Environ. 2022 Feb 10;807(Pt 2):150802. doi: 10.1016/j.scitotenv.2021.150802. Epub 2021 Oct 7. Sci Total Environ. 2022. PMID: 34626628
-
Sub-chronic exposure to antibiotics doxycycline, oxytetracycline or florfenicol impacts gut barrier and induces gut microbiota dysbiosis in adult zebrafish (Daino rerio).Ecotoxicol Environ Saf. 2021 Sep 15;221:112464. doi: 10.1016/j.ecoenv.2021.112464. Epub 2021 Jun 29. Ecotoxicol Environ Saf. 2021. PMID: 34198189
-
Soil oxytetracycline exposure alters the microbial community and enhances the abundance of antibiotic resistance genes in the gut of Enchytraeus crypticus.Sci Total Environ. 2019 Jul 10;673:357-366. doi: 10.1016/j.scitotenv.2019.04.103. Epub 2019 Apr 9. Sci Total Environ. 2019. PMID: 30991325
-
Dose-dependent impact of oxytetracycline on the veal calf microbiome and resistome.BMC Genomics. 2019 Jan 19;20(1):65. doi: 10.1186/s12864-018-5419-x. BMC Genomics. 2019. PMID: 30660184 Free PMC article.
Cited by
-
Effect of Enrofloxacin on the Microbiome, Metabolome, and Abundance of Antibiotic Resistance Genes in the Chicken Cecum.Microbiol Spectr. 2023 Feb 22;11(2):e0479522. doi: 10.1128/spectrum.04795-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36840593 Free PMC article.
-
High prevalence of low-concentration antimicrobial residues in commercial fish: A public health concern in Bangladesh.PLoS One. 2025 May 27;20(5):e0324263. doi: 10.1371/journal.pone.0324263. eCollection 2025. PLoS One. 2025. PMID: 40424403 Free PMC article.
-
Persistent Dysbiosis, Parasite Rise and Growth Impairment in Aquacultured European Seabass after Oxytetracycline Treatment.Microorganisms. 2023 Sep 13;11(9):2302. doi: 10.3390/microorganisms11092302. Microorganisms. 2023. PMID: 37764146 Free PMC article.
References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Bioinformatics, Babraham Institute.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

