Microfluidics combined with fluorescence in situ hybridization (FISH) for Candida spp. detection
- PMID: 36213081
- PMCID: PMC9539416
- DOI: 10.3389/fbioe.2022.987669
Microfluidics combined with fluorescence in situ hybridization (FISH) for Candida spp. detection
Abstract
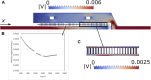
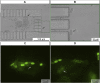
One of the most prevalent healthcare-associated infection is the urinary tract infection (UTI), caused by opportunistic pathogens such as Candida albicans or non-albicans Candida species (NACS). Urine culture methods are routinely used for UTI diagnostics due to their specificity, sensitivity and low-cost. However, these methods are also laborious, time- and reagent-consuming. Therefore, diagnostic methods relying on nucleic acids have been suggested as alternatives. Nucleic acid-based methods can provide results within 24 h and can be adapted to point-of-care (POC) detection. Here, we propose to combine fluorescence in situ hybridization (FISH) with a microfluidic platform for the detection of Candida spp. As a case study we used C. tropicalis, which is reported as the second most common NACS urine isolate obtained from patients suspected with UTI. The microfluidic platform proposed in this study relies on hydrodynamic trapping, and uses physical barriers (e.g., microposts) for the separation of target cells from the suspension. Using a specific peptide nucleic acid (PNA) probe, the FISH procedure was applied onto previously trapped C. tropicalis cells present inside the microfluidic platform. Fluorescence signal intensity of hybridized cells was captured directly under the epifluorescence microscope. Overall, the PNA probe successfully detected C. tropicalis in pure culture and artificial urine (AU) using FISH combined with the microfluidic platform. Our findings reveal that FISH using nucleic acid mimics (PNA) in combination with microfluidics is a reliable method for the detection of microorganisms such as C. tropicalis. As such, this work provides the basis for the development of a POC detection platform in the future.
Keywords: C. tropicalis; FISH; UTI; detection; microfluidics.
Copyright © 2022 Barbosa, Rodrigues, Cerqueira, Miranda and Azevedo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Almeida C., Cerqueira L., Azevedo N. F., Vieira M. J. (2013b). Detection of Salmonella enterica serovar Enteritidis using real time PCR, immunocapture assay, PNA FISH and standard culture methods in different types of food samples. Int. J. Food Microbiol. 161, 16–22. 10.1016/j.ijfoodmicro.2012.11.014 - DOI - PubMed
-
- Almeida C., Sousa J. M., Rocha R., Cerqueira L., Fanning S., Azevedo N. F., et al. (2013c). Detection of Escherichia coli O157 by peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) and comparison to a standard culture method. Appl. Environ. Microbiol. 79, 6293–6300. 10.1128/AEM.01009-13 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

