Requirements for MRN endonuclease processing of topoisomerase II-mediated DNA damage in mammalian cells
- PMID: 36213114
- PMCID: PMC9537633
- DOI: 10.3389/fmolb.2022.1007064
Requirements for MRN endonuclease processing of topoisomerase II-mediated DNA damage in mammalian cells
Abstract
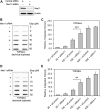
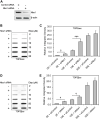
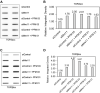
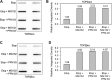
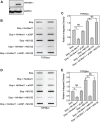
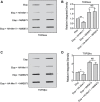
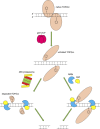
During a normal topoisomerase II (TOP2) reaction, the enzyme forms a covalent enzyme DNA intermediate consisting of a 5' phosphotyrosyl linkage between the enzyme and DNA. While the enzyme typically rejoins the transient breakage after strand passage, a variety of conditions including drugs targeting TOP2 can inhibit DNA resealing, leading to enzyme-mediated DNA damage. A critical aspect of the repair of TOP2-mediated damage is the removal of the TOP2 protein covalently bound to DNA. While proteolysis plays a role in repairing this damage, nucleolytic enzymes must remove the phosphotyrosyl-linked peptide bound to DNA. The MRN complex has been shown to participate in the removal of TOP2 protein from DNA following cellular treatment with TOP2 poisons. In this report we used an optimized ICE (In vivo Complex of Enzyme) assay to measure covalent TOP2/DNA complexes. In agreement with previous independent reports, we find that the absence or inhibition of the MRE11 endonuclease results in elevated levels of both TOP2α and TOP2β covalent complexes. We also examined levels of TOP2 covalent complexes in cells treated with the proteasome inhibitor MG132. Although MRE11 inhibition plus MG132 was not synergistic in etoposide-treated cells, ectopic overexpression of MRE11 resulted in removal of TOP2 even in the presence of MG132. We also found that VCP/p97 inhibition led to elevated TOP2 covalent complexes and prevented the removal of TOP2 covalent complexes by MRE11 overexpression. Our results demonstrate the existence of multiple pathways for proteolytic processing of TOP2 prior to nucleolytic processing, and that MRE11 can process TOP2 covalent complexes even when the proteasome is inhibited. The interactions between VCP/p97 and proteolytic processing of TOP2 covalent complexes merit additional investigation.
Keywords: DNA-protein crosslink (DPC) repair; Mre11-Rad50-Nbs1 complex; VCP/p97; proteasome; topoisomerase II (TOP2).
Copyright © 2022 Sun, Soans, Mishina, Petricci, Pommier, Nitiss and Nitiss.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

