A Methylation Diagnostic Model Based on Random Forests and Neural Networks for Asthma Identification
- PMID: 36213574
- PMCID: PMC9534672
- DOI: 10.1155/2022/2679050
A Methylation Diagnostic Model Based on Random Forests and Neural Networks for Asthma Identification
Abstract
Background: Asthma significantly impacts human life and health as a chronic disease. Traditional treatments for asthma have several limitations. Artificial intelligence aids in cancer treatment and may also accelerate our understanding of asthma mechanisms. We aimed to develop a new clinical diagnosis model for asthma using artificial neural networks (ANN).
Methods: Datasets (GSE85566, GSE40576, and GSE13716) were downloaded from Gene Expression Omnibus (GEO) and identified differentially expressed CpGs (DECs) enriched by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Random forest (RF) and ANN algorithms further identified gene characteristics and built clinical models. In addition, two external validation datasets (GSE40576 and GSE137716) were used to validate the diagnostic ability of the model.
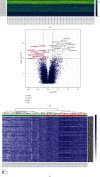
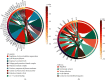
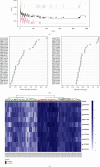
Results: The methylation analysis tool (ChAMP) considered DECs that were up-regulated (n =121) and down-regulated (n =20). GO results showed enrichment of actin cytoskeleton organization and cell-substrate adhesion, shigellosis, and serotonergic synapses. RF (random forest) analysis identified 10 crucial DECs (cg05075579, cg20434422, cg03907390, cg00712106, cg05696969, cg22862094, cg11733958, cg00328720, and cg13570822). ANN constructed the clinical model according to 10 DECs. In two external validation datasets (GSE40576 and GSE137716), the Area Under Curve (AUC) for GSE137716 was 1.000, and AUC for GSE40576 was 0.950, confirming the reliability of the model.
Conclusion: Our findings provide new methylation markers and clinical diagnostic models for asthma diagnosis and treatment.
Copyright © 2022 Dong-Dong Li et al.
Conflict of interest statement
This research does not include any research conducted by any author on human participants or animals. The authors declare no competing interests.
Figures

Similar articles
-
Development and Verification of a Combined Diagnostic Model for Sarcopenia with Random Forest and Artificial Neural Network.Comput Math Methods Med. 2022 Aug 23;2022:2957731. doi: 10.1155/2022/2957731. eCollection 2022. Comput Math Methods Med. 2022. PMID: 36050999 Free PMC article.
-
DNA methylation and gene expression profiles to identify childhood atopic asthma associated genes.BMC Pulm Med. 2021 Sep 15;21(1):292. doi: 10.1186/s12890-021-01655-8. BMC Pulm Med. 2021. PMID: 34525985 Free PMC article.
-
Establishment and Analysis of an Artificial Neural Network Model for Early Detection of Polycystic Ovary Syndrome Using Machine Learning Techniques.J Inflamm Res. 2023 Nov 29;16:5667-5676. doi: 10.2147/JIR.S438838. eCollection 2023. J Inflamm Res. 2023. PMID: 38050562 Free PMC article.
-
Establishment of a Combined Diagnostic Model of Abdominal Aortic Aneurysm with Random Forest and Artificial Neural Network.Biomed Res Int. 2022 Mar 7;2022:7173972. doi: 10.1155/2022/7173972. eCollection 2022. Biomed Res Int. 2022. PMID: 35299890 Free PMC article.
-
Coordinated DNA Methylation and Gene Expression Data for Identification of the Critical Genes Associated with Childhood Atopic Asthma.J Comput Biol. 2020 Jan;27(1):109-120. doi: 10.1089/cmb.2019.0194. Epub 2019 Aug 28. J Comput Biol. 2020. PMID: 31460781
Cited by
-
Development and verification of a combined diagnostic model for primary Sjögren's syndrome by integrated bioinformatics analysis and machine learning.Sci Rep. 2023 May 27;13(1):8641. doi: 10.1038/s41598-023-35864-4. Sci Rep. 2023. PMID: 37244954 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

