Contribution of allergy in the acquisition of uncontrolled severe asthma
- PMID: 36213665
- PMCID: PMC9532527
- DOI: 10.3389/fmed.2022.1009324
Contribution of allergy in the acquisition of uncontrolled severe asthma
Abstract
Asthma is a multifactorial, heterogeneous disease that has a challenging management. It can be divided in non-allergic and allergic (usually associated with house dust mites (HDM) sensitization). There are several treatments options for asthma (corticosteroids, bronchodilators, antileukotrienes, anticholinergics,…); however, there is a subset of patients that do not respond to any of the treatments, who can display either a T2 or a non-T2 phenotype. A deeper understanding of the differential mechanisms underlying each phenotype will help to decipher the contribution of allergy to the acquisition of this uncontrolled severe phenotype. Here, we aim to elucidate the biological pathways associated to allergy in the uncontrolled severe asthmatic phenotype. To do so, twenty-three severe uncontrolled asthmatic patients both with and without HDM-allergy were recruited from Hospital Universitario de Gran Canaria Dr. Negrin. A metabolomic fingerprint was obtained through liquid chromatography coupled to mass spectrometry, and identified metabolites were associated with their pathways. 9/23 patients had uncontrolled HDM-allergic asthma (UCA), whereas 14 had uncontrolled, non-allergic asthma (UCNA). 7/14 (50%) of the UCNA patients had Aspirin Exacerbated Respiratory Disease. There were no significant differences regarding gender or body mass index; but there were significant differences in age and onset age, which were higher in UCNA patients; and in total IgE, which was higher in UCA. The metabolic fingerprint revealed that 103 features were significantly different between UCNA and UCA (p < 0.05), with 97 being increased in UCA and 6 being decreased. We identified lysophosphocholines (LPC) 18:2, 18:3 and 20:4 (increased in UCA patients); and deoxycholic acid and palmitoleoylcarnitine (decreased in UCA). These metabolites were related with a higher activation of phospholipase A2 (PLA2) and other phospholipid metabolism pathways. Our results show that allergy induces the activation of specific inflammatory pathways, such as the PLA2 pathway, which supports its role in the development of an uncontrolled asthma phenotype. There are also clinical differences, such as higher levels of IgE and earlier onset ages for the allergic asthmatic group, as expected. These results provide evidences to better understand the contribution of allergy to the establishment of a severe uncontrolled phenotype.
Keywords: HDM-allergy; allergy; asthma; bile acids (BAs); lysophospholipids; metabolomics.
Copyright © 2022 Delgado Dolset, Obeso, Rodriguez-Coira, Villaseñor, González Cuervo, Arjona, Barbas, Barber, Carrillo and Escribese.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
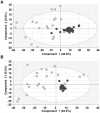
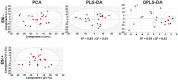


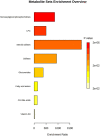
References
-
- Global Initiative for Asthma . Global Strategy for Asthma Management and Prevention. (2022). Available online at: www.ginasthma.org (accessed June 26, 2022).
-
- Vos T, Lim SS, Abbafati C, Abbas KM, Abbasi M, Abbasifard M, et al. . Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. (2020) 396:1204–22. 10.1016/S0140-6736(20)30925-9 - DOI - PMC - PubMed
-
- Campo P, Rodríguez F, Sánchez-García S, Barranco P, Quirce S, Pérez-Francés C, et al. . Phenotypes and endotypes of uncontrolled severe asthma: new treatments. J Investig Allergol Clin Immunol. (2013) 23:76–88. - PubMed
LinkOut - more resources
Full Text Sources

