TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing
- PMID: 36215168
- PMCID: PMC10237422
- DOI: 10.1093/neuonc/noac233
TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing
Abstract
Background: IDH mutant gliomas are grouped into astrocytomas or oligodendrogliomas depending on the codeletion of chromosome arms 1p and 19q. Although the genomic alterations of IDH mutant gliomas have been well described, transcriptional changes unique to either tumor type have not been fully understood. Here, we identify Tripartite Motif Containing 67 (TRIM67), an E3 ubiquitin ligase with essential roles during neuronal development, as an oncogene distinctly upregulated in oligodendrogliomas.
Methods: We used several cell lines, including patient-derived oligodendroglioma tumorspheres, to knock down or overexpress TRIM67. We coupled high-throughput assays, including RNA sequencing, total lysate-mass spectrometry (MS), and coimmunoprecipitation (co-IP)-MS with functional assays including immunofluorescence (IF) staining, co-IP, and western blotting (WB) to assess the in vitro phenotype associated with TRIM67. Patient-derived oligodendroglioma tumorspheres were orthotopically implanted in mice to determine the effect of TRIM67 on tumor growth and survival.
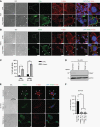
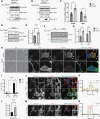
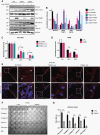
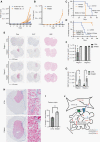
Results: TRIM67 overexpression alters the abundance of cytoskeletal proteins and induces membrane bleb formation. TRIM67-associated blebbing was reverted with the nonmuscle class II myosin inhibitor blebbistatin and selective ROCK inhibitor fasudil. NOGO-A/Rho GTPase/ROCK2 signaling is altered upon TRIM67 ectopic expression, pointing to the underlying mechanism for TRIM67-induced blebbing. Phenotypically, TRIM67 expression resulted in higher cell motility and reduced cell adherence. In orthotopic implantation models of patient-derived oligodendrogliomas, TRIM67 accelerated tumor growth, reduced overall survival, and led to increased vimentin expression at the tumor margin.
Conclusions: Taken together, our results demonstrate that upregulated TRIM67 induces blebbing-based rounded cell morphology through Rho GTPase/ROCK-mediated signaling thereby contributing to glioma pathogenesis.
Keywords: E3 ligase; Rho GTPase signaling; TRIM67; glioma; membrane blebbing; mouse.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Comment in
-
How to move a "fried egg": membrane blebbing in oligodendrogliomas.Neuro Oncol. 2023 Jun 2;25(6):1044-1046. doi: 10.1093/neuonc/noad039. Neuro Oncol. 2023. PMID: 36782080 Free PMC article. No abstract available.
References
-
- Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

