EpiLPS: A fast and flexible Bayesian tool for estimation of the time-varying reproduction number
- PMID: 36215319
- PMCID: PMC9584461
- DOI: 10.1371/journal.pcbi.1010618
EpiLPS: A fast and flexible Bayesian tool for estimation of the time-varying reproduction number
Abstract
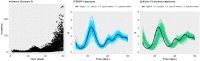
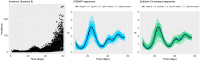
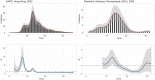
In infectious disease epidemiology, the instantaneous reproduction number [Formula: see text] is a time-varying parameter defined as the average number of secondary infections generated by an infected individual at time t. It is therefore a crucial epidemiological statistic that assists public health decision makers in the management of an epidemic. We present a new Bayesian tool (EpiLPS) for robust estimation of the time-varying reproduction number. The proposed methodology smooths the epidemic curve and allows to obtain (approximate) point estimates and credible intervals of [Formula: see text] by employing the renewal equation, using Bayesian P-splines coupled with Laplace approximations of the conditional posterior of the spline vector. Two alternative approaches for inference are presented: (1) an approach based on a maximum a posteriori argument for the model hyperparameters, delivering estimates of [Formula: see text] in only a few seconds; and (2) an approach based on a Markov chain Monte Carlo (MCMC) scheme with underlying Langevin dynamics for efficient sampling of the posterior target distribution. Case counts per unit of time are assumed to follow a negative binomial distribution to account for potential overdispersion in the data that would not be captured by a classic Poisson model. Furthermore, after smoothing the epidemic curve, a "plug-in'' estimate of the reproduction number can be obtained from the renewal equation yielding a closed form expression of [Formula: see text] as a function of the spline parameters. The approach is extremely fast and free of arbitrary smoothing assumptions. EpiLPS is applied on data of SARS-CoV-1 in Hong-Kong (2003), influenza A H1N1 (2009) in the USA and on the SARS-CoV-2 pandemic (2020-2021) for Belgium, Portugal, Denmark and France.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Cori A. EpiEstim: estimate time varying reproduction numbers from epidemic curves (CRAN); 2021. Available from: https://cran.r-project.org/web/packages/EpiEstim/index.html.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

