Direct observation of stepping rotation of V-ATPase reveals rigid component in coupling between Vo and V1 motors
- PMID: 36215468
- PMCID: PMC9586324
- DOI: 10.1073/pnas.2210204119
Direct observation of stepping rotation of V-ATPase reveals rigid component in coupling between Vo and V1 motors
Abstract
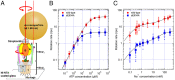
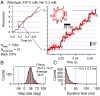
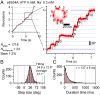
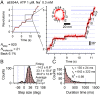
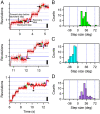
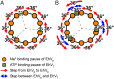
V-ATPases are rotary motor proteins that convert the chemical energy of ATP into the electrochemical potential of ions across cell membranes. V-ATPases consist of two rotary motors, Vo and V1, and Enterococcus hirae V-ATPase (EhVoV1) actively transports Na+ in Vo (EhVo) by using torque generated by ATP hydrolysis in V1 (EhV1). Here, we observed ATP-driven stepping rotation of detergent-solubilized EhVoV1 wild-type, aE634A, and BR350K mutants under various Na+ and ATP concentrations ([Na+] and [ATP], respectively) by using a 40-nm gold nanoparticle as a low-load probe. When [Na+] was low and [ATP] was high, under the condition that only Na+ binding to EhVo is rate limiting, wild-type and aE634A exhibited 10 pausing positions reflecting 10-fold symmetry of the EhVo rotor and almost no backward steps. Duration time before the forward steps was inversely proportional to [Na+], confirming that Na+ binding triggers the steps. When both [ATP] and [Na+] were low, under the condition that both Na+ and ATP bindings are rate limiting, aE634A exhibited 13 pausing positions reflecting 10- and 3-fold symmetries of EhVo and EhV1, respectively. The distribution of duration time before the forward step was fitted well by the sum of two exponential decay functions with distinct time constants. Furthermore, occasional backward steps smaller than 36° were observed. Small backward steps were also observed during three long ATP cleavage pauses of BR350K. These results indicate that EhVo and EhV1 do not share pausing positions, Na+ and ATP bindings occur at different angles, and the coupling between EhVo and EhV1 has a rigid component.
Keywords: V-ATPase; molecular motors; single-molecule analysis.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Torque generation of Enterococcus hirae V-ATPase.J Biol Chem. 2014 Nov 7;289(45):31212-23. doi: 10.1074/jbc.M114.598177. Epub 2014 Sep 25. J Biol Chem. 2014. PMID: 25258315 Free PMC article.
-
Single-molecule analysis reveals rotational substeps and chemo-mechanical coupling scheme of Enterococcus hirae V1-ATPase.J Biol Chem. 2019 Nov 8;294(45):17017-17030. doi: 10.1074/jbc.RA119.008947. Epub 2019 Sep 13. J Biol Chem. 2019. PMID: 31519751 Free PMC article.
-
Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase.J Biol Chem. 2013 Nov 8;288(45):32700-32707. doi: 10.1074/jbc.M113.506329. Epub 2013 Oct 2. J Biol Chem. 2013. PMID: 24089518 Free PMC article.
-
Molecular structure and rotary dynamics of Enterococcus hirae V₁-ATPase.IUBMB Life. 2014 Sep;66(9):624-30. doi: 10.1002/iub.1311. Epub 2014 Sep 17. IUBMB Life. 2014. PMID: 25229752 Review.
-
Structure and dynamics of rotary V1 motor.Cell Mol Life Sci. 2018 May;75(10):1789-1802. doi: 10.1007/s00018-018-2758-3. Epub 2018 Jan 31. Cell Mol Life Sci. 2018. PMID: 29387903 Free PMC article. Review.
Cited by
-
Six states of Enterococcus hirae V-type ATPase reveals non-uniform rotor rotation during turnover.Commun Biol. 2023 Jul 28;6(1):755. doi: 10.1038/s42003-023-05110-8. Commun Biol. 2023. PMID: 37507515 Free PMC article.
-
Tackle "Molecular Engine" by early-career researchers.Biophys Physicobiol. 2022 Sep 22;19:e190039. doi: 10.2142/biophysico.bppb-v19.0039. eCollection 2022. Biophys Physicobiol. 2022. PMID: 36349330 Free PMC article. No abstract available.
-
Visualizing Single V-ATPase Rotation Using Janus Nanoparticles.Nano Lett. 2024 Dec 11;24(49):15638-15644. doi: 10.1021/acs.nanolett.4c04109. Epub 2024 Nov 22. Nano Lett. 2024. PMID: 39573818 Free PMC article.
-
ATP synthesis of Enterococcus hirae V-ATPase driven by sodium motive force.J Biol Chem. 2025 Apr;301(4):108422. doi: 10.1016/j.jbc.2025.108422. Epub 2025 Mar 19. J Biol Chem. 2025. PMID: 40118453 Free PMC article.
-
Visualizing Single V-ATPase Rotation Using Janus Nanoparticles.bioRxiv [Preprint]. 2024 Aug 22:2024.08.22.609254. doi: 10.1101/2024.08.22.609254. bioRxiv. 2024. Update in: Nano Lett. 2024 Dec 11;24(49):15638-15644. doi: 10.1021/acs.nanolett.4c04109. PMID: 39229122 Free PMC article. Updated. Preprint.
References
-
- Yasuda R., Noji H., Yoshida M., Kinosita K. Jr., Itoh H., Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001). - PubMed
-
- Forgac M., Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 (2007). - PubMed
-
- Walker J. E., The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16 (2013). - PubMed
-
- Boyer P. D., The ATP synthase—A splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 (1997). - PubMed
-
- Vasanthakumar T., Rubinstein J. L., Structure and roles of V-type ATPases. Trends Biochem. Sci. 45, 295–307 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

