Origin recognition complex harbors an intrinsic nucleosome remodeling activity
- PMID: 36215487
- PMCID: PMC9586268
- DOI: 10.1073/pnas.2211568119
Origin recognition complex harbors an intrinsic nucleosome remodeling activity
Abstract
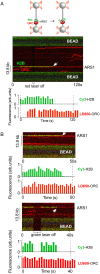
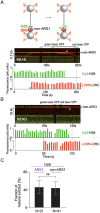
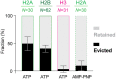
Eukaryotic DNA replication is initiated at multiple chromosomal sites known as origins of replication that are specifically recognized by the origin recognition complex (ORC) containing multiple ATPase sites. In budding yeast, ORC binds to specific DNA sequences known as autonomously replicating sequences (ARSs) that are mostly nucleosome depleted. However, nucleosomes may still inhibit the licensing of some origins by occluding ORC binding and subsequent MCM helicase loading. Using purified proteins and single-molecule visualization, we find here that the ORC can eject histones from a nucleosome in an ATP-dependent manner. The ORC selectively evicts H2A-H2B dimers but leaves the (H3-H4)2 tetramer on DNA. It also discriminates canonical H2A from the H2A.Z variant, evicting the former while retaining the latter. Finally, the bromo-adjacent homology (BAH) domain of the Orc1 subunit is essential for ORC-mediated histone eviction. These findings suggest that the ORC is a bona fide nucleosome remodeler that functions to create a local chromatin environment optimal for origin activity.
Keywords: DNA replication; nucleosome; nucleosome remodeling; origin of replication; origin recognition complex.
Conflict of interest statement
The authors declare no competing interest.
Figures





Similar articles
-
Establishment and function of chromatin organization at replication origins.Nature. 2023 Apr;616(7958):836-842. doi: 10.1038/s41586-023-05926-8. Epub 2023 Apr 5. Nature. 2023. PMID: 37020028
-
Concerted interaction between origin recognition complex (ORC), nucleosomes and replication origin DNA ensures stable ORC-origin binding.Genes Cells. 2013 Sep;18(9):764-79. doi: 10.1111/gtc.12073. Epub 2013 Jun 24. Genes Cells. 2013. PMID: 23795651
-
H2A histone-fold and DNA elements in nucleosome activate SWR1-mediated H2A.Z replacement in budding yeast.Elife. 2015 Jun 27;4:e06845. doi: 10.7554/eLife.06845. Elife. 2015. PMID: 26116819 Free PMC article.
-
Promoters are key organizers of the duplication of vertebrate genomes.Bioessays. 2021 Oct;43(10):e2100141. doi: 10.1002/bies.202100141. Epub 2021 Jul 28. Bioessays. 2021. PMID: 34319621 Review.
-
Molecular Mechanism for Chromatin Regulation During MCM Loading in Mammalian Cells.Adv Exp Med Biol. 2017;1042:61-78. doi: 10.1007/978-981-10-6955-0_3. Adv Exp Med Biol. 2017. PMID: 29357053 Review.
Cited by
-
Chromatin's Influence on Pre-Replication Complex Assembly and Function.Biology (Basel). 2024 Feb 27;13(3):152. doi: 10.3390/biology13030152. Biology (Basel). 2024. PMID: 38534422 Free PMC article. Review.
-
Histone variant H2A.Z cooperates with EBNA1 to maintain Epstein-Barr virus latent epigenome.mBio. 2025 Aug 13;16(8):e0030225. doi: 10.1128/mbio.00302-25. Epub 2025 Jul 14. mBio. 2025. PMID: 40657913 Free PMC article.
-
ORChestra coordinates the replication and repair music.Bioessays. 2023 Apr;45(4):e2200229. doi: 10.1002/bies.202200229. Epub 2023 Feb 22. Bioessays. 2023. PMID: 36811379 Free PMC article.
-
Mechanisms for licensing origins of DNA replication in eukaryotic cells.Nat Struct Mol Biol. 2025 Jul;32(7):1143-1153. doi: 10.1038/s41594-025-01587-5. Epub 2025 Jun 30. Nat Struct Mol Biol. 2025. PMID: 40588663 Review.
-
DNA replication: Mechanisms and therapeutic interventions for diseases.MedComm (2020). 2023 Feb 5;4(1):e210. doi: 10.1002/mco2.210. eCollection 2023 Feb. MedComm (2020). 2023. PMID: 36776764 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01GM118682/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R35GM139654/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R01GM115809/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 GM115809/GM/NIGMS NIH HHS/United States
- R35 GM139654/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

