Tetraspanin-5-mediated MHC class I clustering is required for optimal CD8 T cell activation
- PMID: 36215490
- PMCID: PMC9586303
- DOI: 10.1073/pnas.2122188119
Tetraspanin-5-mediated MHC class I clustering is required for optimal CD8 T cell activation
Abstract
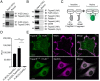
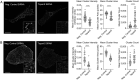
MHC molecules are not randomly distributed on the plasma membrane but instead are present in discrete nanoclusters. The mechanisms that control formation of MHC I nanoclusters and the importance of such structures are incompletely understood. Here, we report a molecular association between tetraspanin-5 (Tspan5) and MHC I molecules that started in the endoplasmic reticulum and was maintained on the plasma membrane. This association was observed both in mouse dendritic cells and in human cancer cell lines. Loss of Tspan5 reduced the size of MHC I clusters without affecting MHC I peptide loading, delivery of complexes to the plasma membrane, or overall surface MHC I levels. Functionally, CD8 T cell responses to antigen presented by Tspan5-deficient dendritic cells were impaired but were restored by antibody-induced reclustering of MHC I molecules. In contrast, Tspan5 did not associate with two other plasma membrane proteins, Flotillin1 and CD55, with or the endoplasmic reticulum proteins Tapasin and TAP. Thus, our findings identify a mechanism underlying the clustering of MHC I molecules that is important for optimal T cell responses.
Keywords: MHC I; Tspan5; antigen presentation; cluster; tetraspanin.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

