Extended longevity geometrically-inverted proximal tubule organoids
- PMID: 36215909
- PMCID: PMC10693433
- DOI: 10.1016/j.biomaterials.2022.121828
Extended longevity geometrically-inverted proximal tubule organoids
Abstract
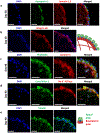
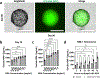
This study reports the cellular self-organization of primary human renal proximal tubule epithelial cells (RPTECs) around a minimal Matrigel scaffold to produce basal-in and apical-out proximal tubule organoids (tubuloids). These tubuloids are produced and maintained in hanging drop cultures for 90+ days, the longest such culture of any kind reported to date. The tubuloids upregulate maturity markers, such as aquaporin-1 (AQP1) and megalin (LRP2), and exhibit less mesenchymal and proliferation markers, such as vimentin and Ki67, compared to 2D cultures. They also experience changes over time as revealed by a comparison of gene expression patterns of cells in 2D culture and in day 31 and day 67 tubuloids. Gene expression analysis and immunohistochemistry reveal an increase in the expression of megalin, an endocytic receptor that can directly bind and uptake protein or potentially assist protein uptake. The tubuloids, including day 90 tubuloids, uptake fluorescent albumin and reveal punctate fluorescent patterns, suggesting functional endocytic uptake through these receptors. Furthermore, the tubuloids release kidney injury molecule-1 (KIM-1), a common biomarker for kidney injury, when exposed to albumin in both dose- and time-dependent manners. While this study focuses on potential applications for modeling proteinuric kidney disease, the tubuloids may have broad utility for studies where apical proximal tubule cell access is required.
Keywords: Basal-in organoid; Kidney; Proteinuria; Tubuloid.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

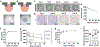


References
-
- King SM, Higgins JW, Nino CR, Smith TR, Paffenroth EH, Fairbairn CE, Docuyanan A, Shah VD, Chen AE, Presnell SC, Nguyen DG, 3D proximal tubule tissues recapitulate key aspects of renal physiology to enable nephrotoxicity testing, Front. Physiol 8 (2017) 1–18. 10.3389/fphys.2017.00123. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

