VEGFR3 modulates brain microvessel branching in a mouse model of 22q11.2 deletion syndrome
- PMID: 36216515
- PMCID: PMC9553901
- DOI: 10.26508/lsa.202101308
VEGFR3 modulates brain microvessel branching in a mouse model of 22q11.2 deletion syndrome
Abstract
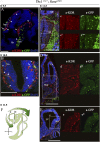
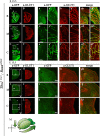
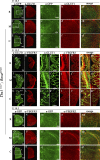
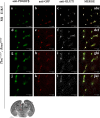
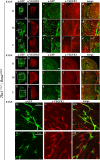
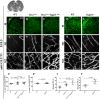
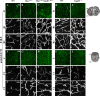
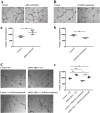
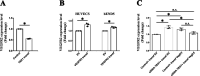
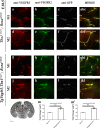
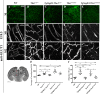
The loss of a single copy of <i>TBX1</i> accounts for most of the clinical signs and symptoms of 22q11.2 deletion syndrome, a common genetic disorder that is characterized by multiple congenital anomalies and brain-related clinical problems, some of which likely have vascular origins. <i>Tbx1</i> mutant mice have brain vascular anomalies, thus making them a useful model to gain insights into the human disease. Here, we found that the main morphogenetic function of TBX1 in the mouse brain is to suppress vessel branching morphogenesis through regulation of <i>Vegfr3</i> We demonstrate that inactivating <i>Vegfr3</i> in the <i>Tbx1</i> expression domain on a <i>Tbx1</i> mutant background enhances brain vessel branching and filopodia formation, whereas increasing <i>Vegfr3</i> expression in this domain fully rescued these phenotypes. Similar results were obtained using an in vitro model of endothelial tubulogenesis. Overall, the results of this study provide genetic evidence that <i>VEGFR3</i> is a regulator of early vessel branching and filopodia formation in the mouse brain and is a likely mediator of the brain vascular phenotype caused by <i>Tbx1</i> loss of function.
© 2022 Cioffi et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
