A high-efficiency scar-free genome-editing toolkit for Acinetobacter baumannii
- PMID: 36216579
- PMCID: PMC9704439
- DOI: 10.1093/jac/dkac328
A high-efficiency scar-free genome-editing toolkit for Acinetobacter baumannii
Abstract
Background: The current mutagenesis tools for Acinetobacter baumannii leave selection markers or residual sequences behind, or involve tedious counterselection and screening steps. Furthermore, they are usually adapted for model strains, rather than for MDR clinical isolates.
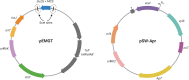
Objectives: To develop a scar-free genome-editing tool suitable for chromosomal and plasmid modifications in MDR A. baumannii AB5075.
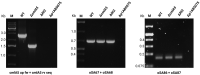
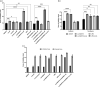
Methods: We prove the efficiency of our adapted genome-editing system by deleting the multidrug efflux pumps craA, cmlA5 and resistance island 2 (RI2), as well as curing plasmid p1AB5075, and combining these mutations. We then characterized the susceptibility of the mutants compared with the WT to different antibiotics (i.e. chloramphenicol, amikacin and tobramycin) by disc diffusion assays and determined the MIC for each strain.
Results: We successfully adapted the genome-editing protocol to A. baumannii AB5075, achieving a double recombination frequency close to 100% and routinely securing the construction of a mutant within 10 working days. Furthermore, we show that both CraA and p1AB5075 are involved in chloramphenicol resistance, and that RI2 and p1AB5075 play a role in resistance to amikacin and tobramycin.
Conclusions: We have developed a versatile and highly efficient genome-editing tool for A. baumannii. We have demonstrated it can be used to modify both the chromosome and native plasmids. By challenging the method, we show the role of CraA and p1AB5075 in antibiotic resistance.
© The Author(s) 2022. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures

References
-
- Dortet L, Legrand P, Soussy CJet al. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J Clin Microbiol 2006; 44: 4471–8. 10.1128/JCM.01535-06 - DOI - PMC - PubMed
-
- McCarthy RR, Larrouy-Maumus GJ, Meiqi Tan MGCet al. Antibiotic resistance mechanisms and their transmission in Acinetobacter baumannii. In: Kishore U, ed. Microbial Pathogenesis: Infection and Immunity. Advances in Experimental Medicine and Biology, 1313. Springer, 2021: 135–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

