The spatial transcriptomic landscape of non-small cell lung cancer brain metastasis
- PMID: 36216799
- PMCID: PMC9551067
- DOI: 10.1038/s41467-022-33365-y
The spatial transcriptomic landscape of non-small cell lung cancer brain metastasis
Abstract
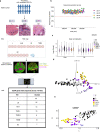
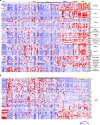
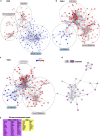
Brain metastases (BrMs) are a common occurrence in lung cancer with a dismal outcome. To understand the mechanism of metastasis to inform prognosis and treatment, here we analyze primary and metastasized tumor specimens from 44 non-small cell lung cancer patients by spatial RNA sequencing, affording a whole transcriptome map of metastasis resolved with morphological markers for the tumor core, tumor immune microenvironment (TIME), and tumor brain microenvironment (TBME). Our data indicate that the tumor microenvironment (TME) in the brain, including the TIME and TBME, undergoes extensive remodeling to create an immunosuppressive and fibrogenic niche for the BrMs. Specifically, the brain TME is characterized with reduced antigen presentation and B/T cell function, increased neutrophils and M2-type macrophages, immature microglia, and reactive astrocytes. Differential gene expression and network analysis identify fibrosis and immune regulation as the major functional modules disrupted in both the lung and brain TME. Besides providing systems-level insights into the mechanism of lung cancer brain metastasis, our study uncovers potential prognostic biomarkers and suggests that therapeutic strategies should be tailored to the immune and fibrosis status of the BrMs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

